DescribePROT: database of amino acid-level protein structure and function predictions
- PMID: 33119734
- PMCID: PMC7778963
- DOI: 10.1093/nar/gkaa931
DescribePROT: database of amino acid-level protein structure and function predictions
Abstract
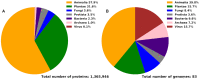
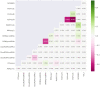
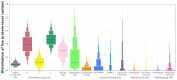
We present DescribePROT, the database of predicted amino acid-level descriptors of structure and function of proteins. DescribePROT delivers a comprehensive collection of 13 complementary descriptors predicted using 10 popular and accurate algorithms for 83 complete proteomes that cover key model organisms. The current version includes 7.8 billion predictions for close to 600 million amino acids in 1.4 million proteins. The descriptors encompass sequence conservation, position specific scoring matrix, secondary structure, solvent accessibility, intrinsic disorder, disordered linkers, signal peptides, MoRFs and interactions with proteins, DNA and RNAs. Users can search DescribePROT by the amino acid sequence and the UniProt accession number and entry name. The pre-computed results are made available instantaneously. The predictions can be accesses via an interactive graphical interface that allows simultaneous analysis of multiple descriptors and can be also downloaded in structured formats at the protein, proteome and whole database scale. The putative annotations included by DescriPROT are useful for a broad range of studies, including: investigations of protein function, applied projects focusing on therapeutics and diseases, and in the development of predictors for other protein sequence descriptors. Future releases will expand the coverage of DescribePROT. DescribePROT can be accessed at http://biomine.cs.vcu.edu/servers/DESCRIBEPROT/.
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

Similar articles
-
DescribePROT in 2023: more, higher-quality and experimental annotations and improved data download options.Nucleic Acids Res. 2024 Jan 5;52(D1):D426-D433. doi: 10.1093/nar/gkad985. Nucleic Acids Res. 2024. PMID: 37933852 Free PMC article.
-
MannDB - a microbial database of automated protein sequence analyses and evidence integration for protein characterization.BMC Bioinformatics. 2006 Oct 17;7:459. doi: 10.1186/1471-2105-7-459. BMC Bioinformatics. 2006. PMID: 17044936 Free PMC article.
-
AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models.Nucleic Acids Res. 2022 Jan 7;50(D1):D439-D444. doi: 10.1093/nar/gkab1061. Nucleic Acids Res. 2022. PMID: 34791371 Free PMC article.
-
Structural protein descriptors in 1-dimension and their sequence-based predictions.Curr Protein Pept Sci. 2011 Sep;12(6):470-89. doi: 10.2174/138920311796957711. Curr Protein Pept Sci. 2011. PMID: 21787299 Review.
-
Predicting Protein-Protein Interactions from the Molecular to the Proteome Level.Chem Rev. 2016 Apr 27;116(8):4884-909. doi: 10.1021/acs.chemrev.5b00683. Epub 2016 Apr 13. Chem Rev. 2016. PMID: 27074302 Review.
Cited by
-
Accurate Prediction of Protein-Binding Residues in Protein Sequences Using SCRIBER.Methods Mol Biol. 2025;2867:247-260. doi: 10.1007/978-1-0716-4196-5_15. Methods Mol Biol. 2025. PMID: 39576586
-
Accurate and Fast Prediction of Intrinsic Disorder Using flDPnn.Methods Mol Biol. 2025;2867:201-218. doi: 10.1007/978-1-0716-4196-5_12. Methods Mol Biol. 2025. PMID: 39576583
-
DescribePROT Database of Residue-Level Protein Structure and Function Annotations.Methods Mol Biol. 2025;2867:169-184. doi: 10.1007/978-1-0716-4196-5_10. Methods Mol Biol. 2025. PMID: 39576581
-
Computational Methods to Investigate Intrinsically Disordered Proteins and their Complexes.ArXiv [Preprint]. 2024 Sep 3:arXiv:2409.02240v1. ArXiv. 2024. PMID: 39279844 Free PMC article. Preprint.
-
Alternate RNA decoding results in stable and abundant proteins in mammals.bioRxiv [Preprint]. 2024 Oct 2:2024.08.26.609665. doi: 10.1101/2024.08.26.609665. bioRxiv. 2024. PMID: 39253435 Free PMC article. Preprint.
References
-
- Boutet E., Lieberherr D., Tognolli M., Schneider M., Bansal P., Bridge A.J., Poux S., Bougueleret L., Xenarios I.. UniProtKB/Swiss-Prot, the manually annotated section of the UniProt KnowledgeBase: how to use the entry view. Methods Mol. Biol. 2016; 1374:23–54. - PubMed
-
- Rost B. Prediction in 1D: secondary structure, membrane helices, and accessibility. Methods Biochem. Anal. 2003; 44:559–587. - PubMed
-
- Kurgan L., Disfani F.M.. Structural protein descriptors in 1-dimension and their sequence-based predictions. Curr. Protein Pept. Sci. 2011; 12:470–489. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources


