Cancer-associated mutations in DICER1 RNase IIIa and IIIb domains exert similar effects on miRNA biogenesis
- PMID: 31417090
- PMCID: PMC6695490
- DOI: 10.1038/s41467-019-11610-1
Cancer-associated mutations in DICER1 RNase IIIa and IIIb domains exert similar effects on miRNA biogenesis
Abstract
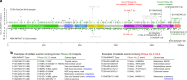
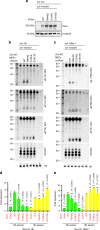
Somatic mutations in the RNase IIIb domain of DICER1 arise in cancer and disrupt the cleavage of 5' pre-miRNA arms. Here, we characterize an unstudied, recurrent, mutation (S1344L) in the DICER1 RNase IIIa domain in tumors from The Cancer Genome Atlas (TCGA) project and MSK-IMPACT profiling. RNase IIIa/b hotspots are absent from most cancers, but are notably enriched in uterine cancers. Systematic analysis of TCGA small RNA datasets show that DICER1 RNase IIIa-S1344L tumors deplete 5p-miRNAs, analogous to RNase IIIb hotspot samples. Structural and evolutionary coupling analyses reveal constrained proximity of RNase IIIa-S1344 to the RNase IIIb catalytic site, rationalizing why mutation of this site phenocopies known hotspot alterations. Finally, examination of DICER1 hotspot endometrial tumors reveals derepression of specific miRNA target signatures. In summary, comprehensive analyses of DICER1 somatic mutations and small RNA data reveal a mechanistic aspect of pre-miRNA processing that manifests in specific cancer settings.
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Recurrent DICER1 hotspot mutations in endometrial tumours and their impact on microRNA biogenesis.J Pathol. 2015 Oct;237(2):215-25. doi: 10.1002/path.4569. Epub 2015 Jul 6. J Pathol. 2015. PMID: 26033159
-
Cancer-associated somatic DICER1 hotspot mutations cause defective miRNA processing and reverse-strand expression bias to predominantly mature 3p strands through loss of 5p strand cleavage.J Pathol. 2013 Feb;229(3):400-9. doi: 10.1002/path.4135. J Pathol. 2013. PMID: 23132766
-
A complex DICER1 syndrome phenotype associated with a germline pathogenic variant affecting the RNase IIIa domain of DICER1.J Med Genet. 2022 Feb;59(2):141-146. doi: 10.1136/jmedgenet-2020-107385. Epub 2020 Nov 18. J Med Genet. 2022. PMID: 33208384 Free PMC article.
-
The co-occurrence of an ovarian Sertoli-Leydig cell tumor with a thyroid carcinoma is highly suggestive of a DICER1 syndrome.Virchows Arch. 2016 May;468(5):631-6. doi: 10.1007/s00428-016-1922-0. Epub 2016 Mar 16. Virchows Arch. 2016. PMID: 26983701 Review.
-
DICER1-associated embryonal rhabdomyosarcoma and adenosarcoma of the gynecologic tract: Pathology, molecular genetics, and indications for molecular testing.Genes Chromosomes Cancer. 2021 Mar;60(3):217-233. doi: 10.1002/gcc.22913. Epub 2020 Nov 19. Genes Chromosomes Cancer. 2021. PMID: 33135284 Review.
Cited by
-
Primary intracranial sarcoma associated with DICER1 mutant: a case report and preclinical investigation.Brain Tumor Pathol. 2024 Nov 10. doi: 10.1007/s10014-024-00495-8. Online ahead of print. Brain Tumor Pathol. 2024. PMID: 39522081
-
Shedding light on the DICER1 mutational spectrum of uncertain significance in malignant neoplasms.Front Mol Biosci. 2024 Oct 3;11:1441180. doi: 10.3389/fmolb.2024.1441180. eCollection 2024. Front Mol Biosci. 2024. PMID: 39421690 Free PMC article.
-
Determinants of selectivity in the dicing mechanism.Nat Commun. 2024 Oct 18;15(1):8989. doi: 10.1038/s41467-024-53322-1. Nat Commun. 2024. PMID: 39420173 Free PMC article.
-
Synthesis and Regulation of miRNA, Its Role in Oncogenesis, and Its Association with Colorectal Cancer Progression, Diagnosis, and Prognosis.Diagnostics (Basel). 2024 Jul 7;14(13):1450. doi: 10.3390/diagnostics14131450. Diagnostics (Basel). 2024. PMID: 39001340 Free PMC article. Review.
-
The microRNA Let-7 and its exosomal form: Epigenetic regulators of gynecological cancers.Cell Biol Toxicol. 2024 Jun 5;40(1):42. doi: 10.1007/s10565-024-09884-3. Cell Biol Toxicol. 2024. PMID: 38836981 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials


