MusiteDeep: a deep-learning framework for general and kinase-specific phosphorylation site prediction
- PMID: 29036382
- PMCID: PMC5860086
- DOI: 10.1093/bioinformatics/btx496
MusiteDeep: a deep-learning framework for general and kinase-specific phosphorylation site prediction
Abstract
Motivation: Computational methods for phosphorylation site prediction play important roles in protein function studies and experimental design. Most existing methods are based on feature extraction, which may result in incomplete or biased features. Deep learning as the cutting-edge machine learning method has the ability to automatically discover complex representations of phosphorylation patterns from the raw sequences, and hence it provides a powerful tool for improvement of phosphorylation site prediction.
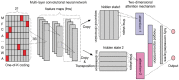
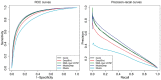
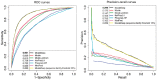
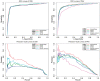
Results: We present MusiteDeep, the first deep-learning framework for predicting general and kinase-specific phosphorylation sites. MusiteDeep takes raw sequence data as input and uses convolutional neural networks with a novel two-dimensional attention mechanism. It achieves over a 50% relative improvement in the area under the precision-recall curve in general phosphorylation site prediction and obtains competitive results in kinase-specific prediction compared to other well-known tools on the benchmark data.
Availability and implementation: MusiteDeep is provided as an open-source tool available at https://github.com/duolinwang/MusiteDeep.
Contact: xudong@missouri.edu.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author 2017. Published by Oxford University Press. All rights reserved. For Permissions, please e-mail: journals.permissions@oup.com
Figures

Similar articles
-
MusiteDeep: a deep-learning based webserver for protein post-translational modification site prediction and visualization.Nucleic Acids Res. 2020 Jul 2;48(W1):W140-W146. doi: 10.1093/nar/gkaa275. Nucleic Acids Res. 2020. PMID: 32324217 Free PMC article.
-
A Pretrained ELECTRA Model for Kinase-Specific Phosphorylation Site Prediction.Methods Mol Biol. 2022;2499:105-124. doi: 10.1007/978-1-0716-2317-6_4. Methods Mol Biol. 2022. PMID: 35696076
-
Capsule network for protein post-translational modification site prediction.Bioinformatics. 2019 Jul 15;35(14):2386-2394. doi: 10.1093/bioinformatics/bty977. Bioinformatics. 2019. PMID: 30520972 Free PMC article.
-
Kinase-specific prediction of protein phosphorylation sites.Methods Mol Biol. 2009;527:299-310, x. doi: 10.1007/978-1-60327-834-8_22. Methods Mol Biol. 2009. PMID: 19241022 Review.
-
A Review of Machine Learning and Algorithmic Methods for Protein Phosphorylation Site Prediction.Genomics Proteomics Bioinformatics. 2023 Dec;21(6):1266-1285. doi: 10.1016/j.gpb.2023.03.007. Epub 2023 Oct 19. Genomics Proteomics Bioinformatics. 2023. PMID: 37863385 Free PMC article. Review.
Cited by
-
GBMPhos: A Gating Mechanism and Bi-GRU-Based Method for Identifying Phosphorylation Sites of SARS-CoV-2 Infection.Biology (Basel). 2024 Oct 6;13(10):798. doi: 10.3390/biology13100798. Biology (Basel). 2024. PMID: 39452107 Free PMC article.
-
Sitetack: a deep learning model that improves PTM prediction by using known PTMs.Bioinformatics. 2024 Nov 1;40(11):btae602. doi: 10.1093/bioinformatics/btae602. Bioinformatics. 2024. PMID: 39388212 Free PMC article.
-
VUStruct: a compute pipeline for high throughput and personalized structural biology.bioRxiv [Preprint]. 2024 Aug 7:2024.08.06.606224. doi: 10.1101/2024.08.06.606224. bioRxiv. 2024. PMID: 39149406 Free PMC article. Preprint.
-
Serotype-Specific Regulation of Dengue Virus NS5 Protein Subcellular Localization.ACS Infect Dis. 2024 Jun 14;10(6):2047-2062. doi: 10.1021/acsinfecdis.4c00054. Epub 2024 May 29. ACS Infect Dis. 2024. PMID: 38811007 Free PMC article.
-
TransPTM: a transformer-based model for non-histone acetylation site prediction.Brief Bioinform. 2024 Mar 27;25(3):bbae219. doi: 10.1093/bib/bbae219. Brief Bioinform. 2024. PMID: 38725156 Free PMC article.
References
-
- Alipanahi B. et al. (2015) Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat. Biotechnol., 33, 831–838. - PubMed
-
- Bahdanau D. et al. (2014) Neural machine translation by jointly learning to align and translate, arXiv preprint arXiv: 1409.0473.
-
- Blom N. et al. (2004) Prediction of post‐translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics, 4, 1633–1649. - PubMed
-
- Caruana R. (1995) Learning many related tasks at the same time with backpropagation. In: Advances in Neural Information Processing Systems, 7, pp. 657–664.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources


