Kingdom Chromista and its eight phyla: a new synthesis emphasising periplastid protein targeting, cytoskeletal and periplastid evolution, and ancient divergences
- PMID: 28875267
- PMCID: PMC5756292
- DOI: 10.1007/s00709-017-1147-3
Kingdom Chromista and its eight phyla: a new synthesis emphasising periplastid protein targeting, cytoskeletal and periplastid evolution, and ancient divergences
Abstract
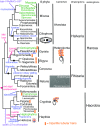
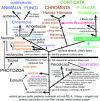
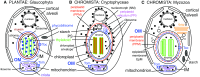
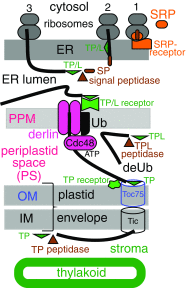
In 1981 I established kingdom Chromista, distinguished from Plantae because of its more complex chloroplast-associated membrane topology and rigid tubular multipartite ciliary hairs. Plantae originated by converting a cyanobacterium to chloroplasts with Toc/Tic translocons; most evolved cell walls early, thereby losing phagotrophy. Chromists originated by enslaving a phagocytosed red alga, surrounding plastids by two extra membranes, placing them within the endomembrane system, necessitating novel protein import machineries. Early chromists retained phagotrophy, remaining naked and repeatedly reverted to heterotrophy by losing chloroplasts. Therefore, Chromista include secondary phagoheterotrophs (notably ciliates, many dinoflagellates, Opalozoa, Rhizaria, heliozoans) or walled osmotrophs (Pseudofungi, Labyrinthulea), formerly considered protozoa or fungi respectively, plus endoparasites (e.g. Sporozoa) and all chromophyte algae (other dinoflagellates, chromeroids, ochrophytes, haptophytes, cryptophytes). I discuss their origin, evolutionary diversification, and reasons for making chromists one kingdom despite highly divergent cytoskeletons and trophic modes, including improved explanations for periplastid/chloroplast protein targeting, derlin evolution, and ciliary/cytoskeletal diversification. I conjecture that transit-peptide-receptor-mediated 'endocytosis' from periplastid membranes generates periplastid vesicles that fuse with the arguably derlin-translocon-containing periplastid reticulum (putative red algal trans-Golgi network homologue; present in all chromophytes except dinoflagellates). I explain chromist origin from ancestral corticates and neokaryotes, reappraising tertiary symbiogenesis; a chromist cytoskeletal synapomorphy, a bypassing microtubule band dextral to both centrioles, favoured multiple axopodial origins. I revise chromist higher classification by transferring rhizarian subphylum Endomyxa from Cercozoa to Retaria; establishing retarian subphylum Ectoreta for Foraminifera plus Radiozoa, apicomonad subclasses, new dinozoan classes Myzodinea (grouping Colpovora gen. n., Psammosa), Endodinea, Sulcodinea, and subclass Karlodinia; and ranking heterokont Gyrista as phylum not superphylum.
Keywords: Chloroplast protein targeting; Chromist evolution; Chromist periplastid membrane; Chromist periplastid reticulum; Microtubular centriolar roots; Sporozoan conoid origin.
Conflict of interest statement
The author declares he has no conflict of interest.
Figures

Similar articles
-
Genomic reduction and evolution of novel genetic membranes and protein-targeting machinery in eukaryote-eukaryote chimaeras (meta-algae).Philos Trans R Soc Lond B Biol Sci. 2003 Jan 29;358(1429):109-33; discussion 133-4. doi: 10.1098/rstb.2002.1194. Philos Trans R Soc Lond B Biol Sci. 2003. PMID: 12594921 Free PMC article.
-
Multigene phylogeny and cell evolution of chromist infrakingdom Rhizaria: contrasting cell organisation of sister phyla Cercozoa and Retaria.Protoplasma. 2018 Sep;255(5):1517-1574. doi: 10.1007/s00709-018-1241-1. Epub 2018 Apr 17. Protoplasma. 2018. PMID: 29666938 Free PMC article.
-
The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa.Int J Syst Evol Microbiol. 2002 Mar;52(Pt 2):297-354. doi: 10.1099/00207713-52-2-297. Int J Syst Evol Microbiol. 2002. PMID: 11931142 Review.
-
Principles of protein and lipid targeting in secondary symbiogenesis: euglenoid, dinoflagellate, and sporozoan plastid origins and the eukaryote family tree.J Eukaryot Microbiol. 1999 Jul-Aug;46(4):347-66. doi: 10.1111/j.1550-7408.1999.tb04614.x. J Eukaryot Microbiol. 1999. PMID: 18092388
-
Ciliary transition zone evolution and the root of the eukaryote tree: implications for opisthokont origin and classification of kingdoms Protozoa, Plantae, and Fungi.Protoplasma. 2022 May;259(3):487-593. doi: 10.1007/s00709-021-01665-7. Epub 2021 Dec 23. Protoplasma. 2022. PMID: 34940909 Free PMC article. Review.
Cited by
-
The evolutionary origins and ancestral features of septins.Front Cell Dev Biol. 2024 Jun 26;12:1406966. doi: 10.3389/fcell.2024.1406966. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 38994454 Free PMC article.
-
The Evolutionary Origins and Ancestral Features of Septins.bioRxiv [Preprint]. 2024 Mar 27:2024.03.25.586683. doi: 10.1101/2024.03.25.586683. bioRxiv. 2024. Update in: Front Cell Dev Biol. 2024 Jun 26;12:1406966. doi: 10.3389/fcell.2024.1406966 PMID: 38585751 Free PMC article. Updated. Preprint.
-
16S and 18S rDNA Amplicon Sequencing Analysis of Aesthetically Problematic Microbial Mats on the Walls of the Petralona Cave: The Use of Essential Oils as a Cleaning Method.Microorganisms. 2023 Oct 31;11(11):2681. doi: 10.3390/microorganisms11112681. Microorganisms. 2023. PMID: 38004693 Free PMC article.
-
Multiple parallel origins of parasitic Marine Alveolates.Nat Commun. 2023 Nov 3;14(1):7049. doi: 10.1038/s41467-023-42807-0. Nat Commun. 2023. PMID: 37923716 Free PMC article.
-
Towards interoperability in infection control: a standard data model for microbiology.Sci Data. 2023 Sep 23;10(1):654. doi: 10.1038/s41597-023-02560-x. Sci Data. 2023. PMID: 37741862 Free PMC article.
References
-
- Amiar S, et al. Apicoplast-localized lysophosphatidic acid precursor assembly is required for bulk phospholipid synthesis in Toxoplasma gondii and relies on an algal/plant-like glycerol 3-phosphate acyltransferase. PLoS Pathog. 2016;12:e1005765. doi: 10.1371/journal.ppat.1005765. - DOI - PMC - PubMed
-
- Anderson OR, Cavalier-Smith T. Ultrastructure of Diplophrys parva, a new small freshwater species, and a revised analysis of Labyrinthulea (Heterokonta) Acta Protozool. 2012;51:291–304.
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources


