PaperBLAST: Text Mining Papers for Information about Homologs
- PMID: 28845458
- PMCID: PMC5557654
- DOI: 10.1128/mSystems.00039-17
PaperBLAST: Text Mining Papers for Information about Homologs
Abstract
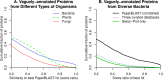
Large-scale genome sequencing has identified millions of protein-coding genes whose function is unknown. Many of these proteins are similar to characterized proteins from other organisms, but much of this information is missing from annotation databases and is hidden in the scientific literature. To make this information accessible, PaperBLAST uses EuropePMC to search the full text of scientific articles for references to genes. PaperBLAST also takes advantage of curated resources (Swiss-Prot, GeneRIF, and EcoCyc) that link protein sequences to scientific articles. PaperBLAST's database includes over 700,000 scientific articles that mention over 400,000 different proteins. Given a protein of interest, PaperBLAST quickly finds similar proteins that are discussed in the literature and presents snippets of text from relevant articles or from the curators. PaperBLAST is available at http://papers.genomics.lbl.gov/. IMPORTANCE With the recent explosion of genome sequencing data, there are now millions of uncharacterized proteins. If a scientist becomes interested in one of these proteins, it can be very difficult to find information as to its likely function. Often a protein whose sequence is similar, and which is likely to have a similar function, has been studied already, but this information is not available in any database. To help find articles about similar proteins, PaperBLAST searches the full text of scientific articles for protein identifiers or gene identifiers, and it links these articles to protein sequences. Then, given a protein of interest, it can quickly find similar proteins in its database by using standard software (BLAST), and it can show snippets of text from relevant papers. We hope that PaperBLAST will make it easier for biologists to predict proteins' functions.
Keywords: annotation; text mining.
Figures

Similar articles
-
Interactive tools for functional annotation of bacterial genomes.Database (Oxford). 2024 Sep 6;2024:baae089. doi: 10.1093/database/baae089. Database (Oxford). 2024. PMID: 39241109 Free PMC article.
-
Curated BLAST for Genomes.mSystems. 2019 Mar 26;4(2):e00072-19. doi: 10.1128/mSystems.00072-19. eCollection 2019 Mar-Apr. mSystems. 2019. PMID: 30944879 Free PMC article.
-
PubMed Text Similarity Model and its application to curation efforts in the Conserved Domain Database.Database (Oxford). 2019 Jan 1;2019:baz064. doi: 10.1093/database/baz064. Database (Oxford). 2019. PMID: 31267135 Free PMC article.
-
A text-mining perspective on the requirements for electronically annotated abstracts.FEBS Lett. 2008 Apr 9;582(8):1178-81. doi: 10.1016/j.febslet.2008.02.072. Epub 2008 Mar 6. FEBS Lett. 2008. PMID: 18328824 Review.
-
Mining biological networks from full-text articles.Methods Mol Biol. 2014;1159:135-45. doi: 10.1007/978-1-4939-0709-0_8. Methods Mol Biol. 2014. PMID: 24788265 Review.
Cited by
-
In vivo manipulation of human gut Bacteroides fitness by abiotic oligosaccharides.Nat Chem Biol. 2024 Oct 23. doi: 10.1038/s41589-024-01763-6. Online ahead of print. Nat Chem Biol. 2024. PMID: 39443715
-
High-throughput protein characterization by complementation using DNA barcoded fragment libraries.Mol Syst Biol. 2024 Nov;20(11):1207-1229. doi: 10.1038/s44320-024-00068-z. Epub 2024 Oct 7. Mol Syst Biol. 2024. PMID: 39375541 Free PMC article.
-
Interactive tools for functional annotation of bacterial genomes.Database (Oxford). 2024 Sep 6;2024:baae089. doi: 10.1093/database/baae089. Database (Oxford). 2024. PMID: 39241109 Free PMC article.
-
Comparative transcriptomics reveals a highly polymorphic Xanthomonas HrpG virulence regulon.BMC Genomics. 2024 Aug 9;25(1):777. doi: 10.1186/s12864-024-10684-6. BMC Genomics. 2024. PMID: 39123115 Free PMC article.
-
Barcoded overexpression screens in gut Bacteroidales identify genes with roles in carbon utilization and stress resistance.Nat Commun. 2024 Aug 5;15(1):6618. doi: 10.1038/s41467-024-50124-3. Nat Commun. 2024. PMID: 39103350 Free PMC article.
References
-
- Poux S, Arighi CN, Magrane M, Bateman A, Wei C-H, Lu Z, Boutet E, Bye-A-Jee H, Famiglietti ML, Roechert B. 2016. On expert curation and sustainability: UniProtKB/Swiss-Prot as a case study. bioRxiv https://doi.org/10.1101/094011. - DOI - PMC - PubMed
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials


