Properties of Gluten Intolerance: Gluten Structure, Evolution, Pathogenicity and Detoxification Capabilities
- PMID: 27763541
- PMCID: PMC5084031
- DOI: 10.3390/nu8100644
Properties of Gluten Intolerance: Gluten Structure, Evolution, Pathogenicity and Detoxification Capabilities
Abstract
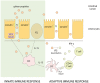
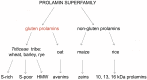
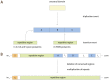
Theterm gluten intolerance may refer to three types of human disorders: autoimmune celiac disease (CD), allergy to wheat and non-celiac gluten sensitivity (NCGS). Gluten is a mixture of prolamin proteins present mostly in wheat, but also in barley, rye and oat. Gluten can be subdivided into three major groups: S-rich, S-poor and high molecular weight proteins. Prolamins within the groups possess similar structures and properties. All gluten proteins are evolutionarily connected and share the same ancestral origin. Gluten proteins are highly resistant to hydrolysis mediated by proteases of the human gastrointestinal tract. It results in emergence of pathogenic peptides, which cause CD and allergy in genetically predisposed people. There is a hierarchy of peptide toxicity and peptide recognition by T cells. Nowadays, there are several ways to detoxify gluten peptides: the most common is gluten-free diet (GFD), which has proved its effectiveness; prevention programs, enzymatic therapy, correction of gluten pathogenicity pathways and genetically modified grains with reduced immunotoxicity. A deep understanding of gluten intolerance underlying mechanisms and detailed knowledge of gluten properties may lead to the emergence of novel effective approaches for treatment of gluten-related disorders.
Keywords: NCGS; avenin; celiac disease; gliadin; gluten; gluten intolerance; glutenin; hordein; secalin; wheat allergy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

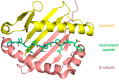
Similar articles
-
[A review of diseases related to the intake of gluten].Nutr Hosp. 2015 Jun 1;31(6):2359-71. doi: 10.3305/nh.2015.31.6.8984. Nutr Hosp. 2015. PMID: 26040340 Review. Spanish.
-
Gluten sensitivities and the allergist: Threshing the grain from the husks.Allergy. 2018 Jul;73(7):1359-1368. doi: 10.1111/all.13354. Epub 2017 Nov 29. Allergy. 2018. PMID: 29131356 Review.
-
Prospects of Developing Medicinal Therapeutic Strategies and Pharmaceutical Design for Effective Gluten Intolerance Treatment.Curr Pharm Des. 2016;22(16):2439-49. doi: 10.2174/1381612822666160201115543. Curr Pharm Des. 2016. PMID: 26831462 Review.
-
Non coeliac gluten sensitivity - A new disease with gluten intolerance.Clin Nutr. 2015 Apr;34(2):189-94. doi: 10.1016/j.clnu.2014.08.012. Epub 2014 Aug 29. Clin Nutr. 2015. PMID: 25245857 Review.
-
Preparation and characterization of enzymatically hydrolyzed prolamins from wheat, rye, and barley as references for the immunochemical quantitation of partially hydrolyzed gluten.Anal Bioanal Chem. 2009 Nov;395(6):1721-8. doi: 10.1007/s00216-009-3080-6. Anal Bioanal Chem. 2009. PMID: 19763549
Cited by
-
Grain albumin content is affected by 1BL/1RS translocation and alters the processing quality of wheat (Triticum aestivum L.).Front Plant Sci. 2024 Jul 22;15:1449826. doi: 10.3389/fpls.2024.1449826. eCollection 2024. Front Plant Sci. 2024. PMID: 39109063 Free PMC article.
-
A comparative study of sericin and gluten for magnetic nanoparticle-mediated drug delivery to breast cancer cell lines.Sci Rep. 2024 Aug 5;14(1):18150. doi: 10.1038/s41598-024-69009-y. Sci Rep. 2024. PMID: 39103485 Free PMC article.
-
Reducing Immunoreactivity of Gluten Peptides by Probiotic Lactic Acid Bacteria for Dietary Management of Gluten-Related Diseases.Nutrients. 2024 Mar 27;16(7):976. doi: 10.3390/nu16070976. Nutrients. 2024. PMID: 38613010 Free PMC article.
-
Celiac Disease: Risks of Cross-Contamination and Strategies for Gluten Removal in Food Environments.Int J Environ Res Public Health. 2024 Jan 24;21(2):124. doi: 10.3390/ijerph21020124. Int J Environ Res Public Health. 2024. PMID: 38397615 Free PMC article. Review.
-
Identification and Growth Characteristics of a Gluten-Degrading Bacterium from Wheat Grains for Gluten-Degrading Enzyme Production.Microorganisms. 2023 Nov 29;11(12):2884. doi: 10.3390/microorganisms11122884. Microorganisms. 2023. PMID: 38138028 Free PMC article.
References
-
- Husby S., Koletzko S., Korponay-Szabo I.R., Mearin M.L., Phillips A., Shamir R., Troncone R., Giersiepen K., Branski D., Catassi C., et al. European Society for Pediatric Gastroenterology, Hepatology, and Nutrition guidelines for the diagnosis of coeliac disease. J. Pediatr. Gastroenterol. Nutr. 2012;54:136–160. doi: 10.1097/MPG.0b013e31821a23d0. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical


