Contribution of Sequence Motif, Chromatin State, and DNA Structure Features to Predictive Models of Transcription Factor Binding in Yeast
- PMID: 26291518
- PMCID: PMC4546298
- DOI: 10.1371/journal.pcbi.1004418
Contribution of Sequence Motif, Chromatin State, and DNA Structure Features to Predictive Models of Transcription Factor Binding in Yeast
Abstract
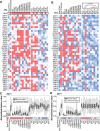
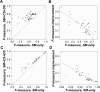
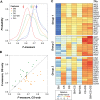
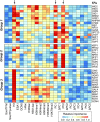
Transcription factor (TF) binding is determined by the presence of specific sequence motifs (SM) and chromatin accessibility, where the latter is influenced by both chromatin state (CS) and DNA structure (DS) properties. Although SM, CS, and DS have been used to predict TF binding sites, a predictive model that jointly considers CS and DS has not been developed to predict either TF-specific binding or general binding properties of TFs. Using budding yeast as model, we found that machine learning classifiers trained with either CS or DS features alone perform better in predicting TF-specific binding compared to SM-based classifiers. In addition, simultaneously considering CS and DS further improves the accuracy of the TF binding predictions, indicating the highly complementary nature of these two properties. The contributions of SM, CS, and DS features to binding site predictions differ greatly between TFs, allowing TF-specific predictions and potentially reflecting different TF binding mechanisms. In addition, a "TF-agnostic" predictive model based on three DNA "intrinsic properties" (in silico predicted nucleosome occupancy, major groove geometry, and dinucleotide free energy) that can be calculated from genomic sequences alone has performance that rivals the model incorporating experiment-derived data. This intrinsic property model allows prediction of binding regions not only across TFs, but also across DNA-binding domain families with distinct structural folds. Furthermore, these predicted binding regions can help identify TF binding sites that have a significant impact on target gene expression. Because the intrinsic property model allows prediction of binding regions across DNA-binding domain families, it is TF agnostic and likely describes general binding potential of TFs. Thus, our findings suggest that it is feasible to establish a TF agnostic model for identifying functional regulatory regions in potentially any sequenced genome.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Assessing the model transferability for prediction of transcription factor binding sites based on chromatin accessibility.BMC Bioinformatics. 2017 Jul 27;18(1):355. doi: 10.1186/s12859-017-1769-7. BMC Bioinformatics. 2017. PMID: 28750606 Free PMC article.
-
Nucleosome free regions in yeast promoters result from competitive binding of transcription factors that interact with chromatin modifiers.PLoS Comput Biol. 2013;9(8):e1003181. doi: 10.1371/journal.pcbi.1003181. Epub 2013 Aug 22. PLoS Comput Biol. 2013. PMID: 23990766 Free PMC article.
-
Predicting transcription factor site occupancy using DNA sequence intrinsic and cell-type specific chromatin features.BMC Bioinformatics. 2016 Jan 11;17 Suppl 1(Suppl 1):4. doi: 10.1186/s12859-015-0846-z. BMC Bioinformatics. 2016. PMID: 26818008 Free PMC article.
-
Mechanisms by which transcription factors gain access to target sequence elements in chromatin.Curr Opin Genet Dev. 2013 Apr;23(2):116-23. doi: 10.1016/j.gde.2012.11.008. Epub 2012 Dec 19. Curr Opin Genet Dev. 2013. PMID: 23266217 Free PMC article. Review.
-
Transcription factor-DNA binding: beyond binding site motifs.Curr Opin Genet Dev. 2017 Apr;43:110-119. doi: 10.1016/j.gde.2017.02.007. Epub 2017 Mar 27. Curr Opin Genet Dev. 2017. PMID: 28359978 Free PMC article. Review.
Cited by
-
Plant Promoters and Terminators for High-Precision Bioengineering.Biodes Res. 2023 Jul 7;5:0013. doi: 10.34133/bdr.0013. eCollection 2023. Biodes Res. 2023. PMID: 37849460 Free PMC article. Review.
-
TSPTFBS 2.0: trans-species prediction of transcription factor binding sites and identification of their core motifs in plants.Front Plant Sci. 2023 May 9;14:1175837. doi: 10.3389/fpls.2023.1175837. eCollection 2023. Front Plant Sci. 2023. PMID: 37229121 Free PMC article.
-
An Evidence Theory and Fuzzy Logic Combined Approach for the Prediction of Potential ARF-Regulated Genes in Quinoa.Plants (Basel). 2022 Dec 23;12(1):71. doi: 10.3390/plants12010071. Plants (Basel). 2022. PMID: 36616201 Free PMC article.
-
Mysteries of gene regulation: Promoters are not the sole triggers of gene expression.Comput Struct Biotechnol J. 2022 Sep 5;20:4910-4920. doi: 10.1016/j.csbj.2022.08.058. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 36147678 Free PMC article.
-
Learning the Regulatory Code of Gene Expression.Front Mol Biosci. 2021 Jun 10;8:673363. doi: 10.3389/fmolb.2021.673363. eCollection 2021. Front Mol Biosci. 2021. PMID: 34179082 Free PMC article. Review.
References
-
- Tompa M, Li N, Bailey TL, Church GM, De Moor B, Eskin E, et al. Assessing computational tools for the discovery of transcription factor binding sites. Nat Biotechnol. 2005;23: 137–44. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous


