Roary: rapid large-scale prokaryote pan genome analysis
- PMID: 26198102
- PMCID: PMC4817141
- DOI: 10.1093/bioinformatics/btv421
Roary: rapid large-scale prokaryote pan genome analysis
Abstract
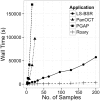
A typical prokaryote population sequencing study can now consist of hundreds or thousands of isolates. Interrogating these datasets can provide detailed insights into the genetic structure of prokaryotic genomes. We introduce Roary, a tool that rapidly builds large-scale pan genomes, identifying the core and accessory genes. Roary makes construction of the pan genome of thousands of prokaryote samples possible on a standard desktop without compromising on the accuracy of results. Using a single CPU Roary can produce a pan genome consisting of 1000 isolates in 4.5 hours using 13 GB of RAM, with further speedups possible using multiple processors.
Availability and implementation: Roary is implemented in Perl and is freely available under an open source GPLv3 license from http://sanger-pathogens.github.io/Roary
Contact: roary@sanger.ac.uk
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author 2015. Published by Oxford University Press.
Figures
Similar articles
-
Large-scale comparative analysis of microbial pan-genomes using PanOCT.Bioinformatics. 2019 Mar 15;35(6):1049-1050. doi: 10.1093/bioinformatics/bty744. Bioinformatics. 2019. PMID: 30165579 Free PMC article.
-
PGAP: pan-genomes analysis pipeline.Bioinformatics. 2012 Feb 1;28(3):416-8. doi: 10.1093/bioinformatics/btr655. Epub 2011 Nov 29. Bioinformatics. 2012. PMID: 22130594 Free PMC article.
-
Prokka: rapid prokaryotic genome annotation.Bioinformatics. 2014 Jul 15;30(14):2068-9. doi: 10.1093/bioinformatics/btu153. Epub 2014 Mar 18. Bioinformatics. 2014. PMID: 24642063
-
NGSPanPipe: A Pipeline for Pan-genome Identification in Microbial Strains from Experimental Reads.Adv Exp Med Biol. 2018;1052:39-49. doi: 10.1007/978-981-10-7572-8_4. Adv Exp Med Biol. 2018. PMID: 29785479 Review.
-
Pan-Genome Storage and Analysis Techniques.Methods Mol Biol. 2018;1704:29-53. doi: 10.1007/978-1-4939-7463-4_2. Methods Mol Biol. 2018. PMID: 29277862 Review.
Cited by
-
Complete genome of single locus sequence typing D1 strain Cutibacterium acnes CN6 isolated from healthy facial skin.BMC Genom Data. 2024 Nov 5;25(1):94. doi: 10.1186/s12863-024-01277-z. BMC Genom Data. 2024. PMID: 39501144
-
Characteristics of Oral Acinetobacter spp. and Evolution of Plasmid-Mediated Carbapenem Resistance in Bacteremia Patients with Hematological Malignancies.Infect Drug Resist. 2024 Oct 30;17:4753-4761. doi: 10.2147/IDR.S478362. eCollection 2024. Infect Drug Resist. 2024. PMID: 39494231 Free PMC article.
-
Comparative genomic analysis and characterization of novel high-quality draft genomes from the coal metagenome.World J Microbiol Biotechnol. 2024 Nov 1;40(12):370. doi: 10.1007/s11274-024-04174-w. World J Microbiol Biotechnol. 2024. PMID: 39485561
-
A Longitudinal Study of Escherichia coli Clinical Isolates from the Tracheal Aspirates of a Paediatric Patient-Strain Type Similar to Pandemic ST131.Microorganisms. 2024 Sep 30;12(10):1990. doi: 10.3390/microorganisms12101990. Microorganisms. 2024. PMID: 39458299 Free PMC article.
-
Investigating Anthrax-Associated Virulence Genes among Archival and Contemporary Bacillus cereus Group Genomes.Pathogens. 2024 Oct 10;13(10):884. doi: 10.3390/pathogens13100884. Pathogens. 2024. PMID: 39452755 Free PMC article.
References
-
- Medini D., et al. (2005) The microbial pan-genome. Curr. Opin. Genet. Dev., 15, 589–594. - PubMed
-
- Nguyen N., et al. (2014) Building a pangenome reference for a population. In: Sharan R. (ed.) Research in Computational Molecular Biology, Springer International Publishing, pp. 207–221.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources