ASTRAL-II: coalescent-based species tree estimation with many hundreds of taxa and thousands of genes
- PMID: 26072508
- PMCID: PMC4765870
- DOI: 10.1093/bioinformatics/btv234
ASTRAL-II: coalescent-based species tree estimation with many hundreds of taxa and thousands of genes
Abstract
Motivation: The estimation of species phylogenies requires multiple loci, since different loci can have different trees due to incomplete lineage sorting, modeled by the multi-species coalescent model. We recently developed a coalescent-based method, ASTRAL, which is statistically consistent under the multi-species coalescent model and which is more accurate than other coalescent-based methods on the datasets we examined. ASTRAL runs in polynomial time, by constraining the search space using a set of allowed 'bipartitions'. Despite the limitation to allowed bipartitions, ASTRAL is statistically consistent.
Results: We present a new version of ASTRAL, which we call ASTRAL-II. We show that ASTRAL-II has substantial advantages over ASTRAL: it is faster, can analyze much larger datasets (up to 1000 species and 1000 genes) and has substantially better accuracy under some conditions. ASTRAL's running time is [Formula: see text], and ASTRAL-II's running time is [Formula: see text], where n is the number of species, k is the number of loci and X is the set of allowed bipartitions for the search space.
Availability and implementation: ASTRAL-II is available in open source at https://github.com/smirarab/ASTRAL and datasets used are available at http://www.cs.utexas.edu/~phylo/datasets/astral2/.
Contact: smirarab@gmail.com
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author 2015. Published by Oxford University Press.
Figures
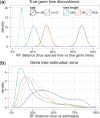
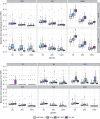
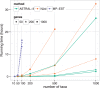
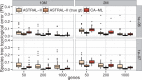

Similar articles
-
ASTRAL: genome-scale coalescent-based species tree estimation.Bioinformatics. 2014 Sep 1;30(17):i541-8. doi: 10.1093/bioinformatics/btu462. Bioinformatics. 2014. PMID: 25161245 Free PMC article.
-
FASTRAL: improving scalability of phylogenomic analysis.Bioinformatics. 2021 Aug 25;37(16):2317-2324. doi: 10.1093/bioinformatics/btab093. Bioinformatics. 2021. PMID: 33576396 Free PMC article.
-
ASTRAL-III: polynomial time species tree reconstruction from partially resolved gene trees.BMC Bioinformatics. 2018 May 8;19(Suppl 6):153. doi: 10.1186/s12859-018-2129-y. BMC Bioinformatics. 2018. PMID: 29745866 Free PMC article.
-
ASTRID: Accurate Species TRees from Internode Distances.BMC Genomics. 2015;16 Suppl 10(Suppl 10):S3. doi: 10.1186/1471-2164-16-S10-S3. Epub 2015 Oct 2. BMC Genomics. 2015. PMID: 26449326 Free PMC article.
-
Approaching Long Genomic Regions and Large Recombination Rates with msParSm as an Alternative to MaCS.Evol Bioinform Online. 2016 Oct 2;12:223-228. doi: 10.4137/EBO.S40268. eCollection 2016. Evol Bioinform Online. 2016. PMID: 27721650 Free PMC article. Review.
Cited by
-
Genome evolution and diversity of wild and cultivated rice species.Nat Commun. 2024 Nov 18;15(1):9994. doi: 10.1038/s41467-024-54427-3. Nat Commun. 2024. PMID: 39557856 Free PMC article.
-
The Meaning and Measure of Concordance Factors in Phylogenomics.Mol Biol Evol. 2024 Nov 1;41(11):msae214. doi: 10.1093/molbev/msae214. Mol Biol Evol. 2024. PMID: 39418118 Free PMC article. Review.
-
A time-calibrated phylogeny of the diversification of Holoadeninae frogs.Front Bioinform. 2024 Oct 2;4:1441373. doi: 10.3389/fbinf.2024.1441373. eCollection 2024. Front Bioinform. 2024. PMID: 39415894 Free PMC article.
-
Short branch attraction in phylogenomic inference under the multispecies coalescent.Front Ecol Evol. 2023;11:1134764. doi: 10.3389/fevo.2023.1134764. Epub 2023 Jun 28. Front Ecol Evol. 2023. PMID: 39233780 Free PMC article.
-
The gradual establishment of complex coumarin biosynthetic pathway in Apiaceae.Nat Commun. 2024 Aug 10;15(1):6864. doi: 10.1038/s41467-024-51285-x. Nat Commun. 2024. PMID: 39127760 Free PMC article.
References
-
- Degnan J.H., Rosenberg N.A. (2009) Gene tree discordance, phylogenetic inference and the multispecies coalescent. Trends Ecol. Evol. , 24, 332–340. - PubMed
-
- Drew B.T., et al. (2014) Another look at the root of the angiosperms reveals a familiar tale. Syst. Biol. , 63, 368–382. - PubMed
-
- Edwards S.V. (2009) Is a new and general theory of molecular systematics emerging? Evolution , 63, 1–19. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous


