Origin of the HIV-1 group O epidemic in western lowland gorillas
- PMID: 25733890
- PMCID: PMC4371950
- DOI: 10.1073/pnas.1502022112
Origin of the HIV-1 group O epidemic in western lowland gorillas
Abstract
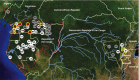
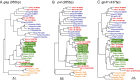
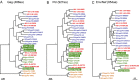
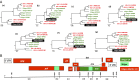
HIV-1, the cause of AIDS, is composed of four phylogenetic lineages, groups M, N, O, and P, each of which resulted from an independent cross-species transmission event of simian immunodeficiency viruses (SIVs) infecting African apes. Although groups M and N have been traced to geographically distinct chimpanzee communities in southern Cameroon, the reservoirs of groups O and P remain unknown. Here, we screened fecal samples from western lowland (n = 2,611), eastern lowland (n = 103), and mountain (n = 218) gorillas for gorilla SIV (SIVgor) antibodies and nucleic acids. Despite testing wild troops throughout southern Cameroon (n = 14), northern Gabon (n = 16), the Democratic Republic of Congo (n = 2), and Uganda (n = 1), SIVgor was identified at only four sites in southern Cameroon, with prevalences ranging from 0.8-22%. Amplification of partial and full-length SIVgor sequences revealed extensive genetic diversity, but all SIVgor strains were derived from a single lineage within the chimpanzee SIV (SIVcpz) radiation. Two fully sequenced gorilla viruses from southwestern Cameroon were very closely related to, and likely represent the source population of, HIV-1 group P. Most of the genome of a third SIVgor strain, from central Cameroon, was very closely related to HIV-1 group O, again pointing to gorillas as the immediate source. Functional analyses identified the cytidine deaminase APOBEC3G as a barrier for chimpanzee-to-gorilla, but not gorilla-to-human, virus transmission. These data indicate that HIV-1 group O, which spreads epidemically in west central Africa and is estimated to have infected around 100,000 people, originated by cross-species transmission from western lowland gorillas.
Keywords: AIDS; HIV-1; SIVgor; gorilla; zoonotic transmission.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Noninvasive follow-up of simian immunodeficiency virus infection in wild-living nonhabituated western lowland gorillas in Cameroon.J Virol. 2012 Sep;86(18):9760-72. doi: 10.1128/JVI.01186-12. Epub 2012 Jun 27. J Virol. 2012. PMID: 22740419 Free PMC article.
-
Origin and biology of simian immunodeficiency virus in wild-living western gorillas.J Virol. 2009 Feb;83(4):1635-48. doi: 10.1128/JVI.02311-08. Epub 2008 Dec 10. J Virol. 2009. PMID: 19073717 Free PMC article.
-
Molecular epidemiology of simian immunodeficiency virus infection in wild-living gorillas.J Virol. 2010 Feb;84(3):1464-76. doi: 10.1128/JVI.02129-09. Epub 2009 Nov 11. J Virol. 2010. PMID: 19906908 Free PMC article.
-
HIV-1 Group O Origin, Evolution, Pathogenesis, and Treatment: Unraveling the Complexity of an Outlier 25 Years Later.AIDS Rev. 2015 Jul-Sep;17(3):147-58. AIDS Rev. 2015. PMID: 26450803 Review.
-
AIDS as a zoonosis? Confusion over the origin of the virus and the origin of the epidemics.J Med Primatol. 2004 Oct;33(5-6):220-6. doi: 10.1111/j.1600-0684.2004.00078.x. J Med Primatol. 2004. PMID: 15525322 Review.
Cited by
-
New Virus Variant Detection Based on the Optimal Natural Metric.Genes (Basel). 2024 Jul 7;15(7):891. doi: 10.3390/genes15070891. Genes (Basel). 2024. PMID: 39062670 Free PMC article.
-
Replicative fitness and pathogenicity of primate lentiviruses in lymphoid tissue, primary human and chimpanzee cells: relation to possible jumps to humans.EBioMedicine. 2024 Feb;100:104965. doi: 10.1016/j.ebiom.2023.104965. Epub 2024 Jan 12. EBioMedicine. 2024. PMID: 38215691 Free PMC article.
-
The HIV-2 OGH double reporter virus shows that HIV-2 is less cytotoxic and less sensitive to reactivation from latency than HIV-1 in cell culture.J Virus Erad. 2023 Aug 29;9(3):100343. doi: 10.1016/j.jve.2023.100343. eCollection 2023 Sep. J Virus Erad. 2023. PMID: 37701289 Free PMC article.
-
A snapshot on HIV-1 evolution through the identification of phylogenetic-specific properties of HIV-1 integrases M/O.PLoS Pathog. 2023 Mar 30;19(3):e1011207. doi: 10.1371/journal.ppat.1011207. eCollection 2023 Mar. PLoS Pathog. 2023. PMID: 36996029 Free PMC article.
-
HIV-1 Non-Group M Strains and ART.Viruses. 2023 Mar 17;15(3):780. doi: 10.3390/v15030780. Viruses. 2023. PMID: 36992488 Free PMC article. Review.
References
-
- Locatelli S, Peeters M. Cross-species transmission of simian retroviruses: How and why they could lead to the emergence of new diseases in the human population. AIDS. 2012;26(6):659–673. - PubMed
-
- Bailes E, et al. Hybrid origin of SIV in chimpanzees. Science. 2003;300(5626):1713. - PubMed
-
- Van Heuverswyn F, et al. Genetic diversity and phylogeographic clustering of SIVcpzPtt in wild chimpanzees in Cameroon. Virology. 2007;368(1):155–171. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
- R37 AI050529/AI/NIAID NIH HHS/United States
- P30 AI045008/AI/NIAID NIH HHS/United States
- R37 AI066998/AI/NIAID NIH HHS/United States
- R37 AI50529/AI/NIAID NIH HHS/United States
- 095831/Wellcome Trust/United Kingdom
- R01 AI 058715/AI/NIAID NIH HHS/United States
- R01 AI064001/AI/NIAID NIH HHS/United States
- R01 AI058715/AI/NIAID NIH HHS/United States
- AI 089246/AI/NIAID NIH HHS/United States
- R01 AI089246/AI/NIAID NIH HHS/United States
- P30 AI 045008/AI/NIAID NIH HHS/United States
- R37 AI 066998/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases


