Conflicting phylogenies for early land plants are caused by composition biases among synonymous substitutions
- PMID: 24399481
- PMCID: PMC3926305
- DOI: 10.1093/sysbio/syt109
Conflicting phylogenies for early land plants are caused by composition biases among synonymous substitutions
Figures

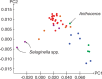
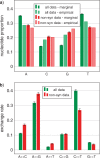
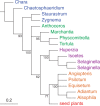
Similar articles
-
Mitochondrial phylogenomics of early land plants: mitigating the effects of saturation, compositional heterogeneity, and codon-usage bias.Syst Biol. 2014 Nov;63(6):862-78. doi: 10.1093/sysbio/syu049. Epub 2014 Jul 28. Syst Biol. 2014. PMID: 25070972
-
Origin of land plants using the multispecies coalescent model.Trends Plant Sci. 2013 Sep;18(9):492-5. doi: 10.1016/j.tplants.2013.04.009. Epub 2013 May 24. Trends Plant Sci. 2013. PMID: 23707196 Review.
-
Codon usage bias and determining forces in green plant mitochondrial genomes.J Integr Plant Biol. 2011 Apr;53(4):324-34. doi: 10.1111/j.1744-7909.2011.01033.x. Epub 2011 Mar 21. J Integr Plant Biol. 2011. PMID: 21332641
-
Complex mutation and weak selection together determined the codon usage bias in bryophyte mitochondrial genomes.J Integr Plant Biol. 2010 Dec;52(12):1100-8. doi: 10.1111/j.1744-7909.2010.00998.x. Epub 2010 Oct 22. J Integr Plant Biol. 2010. PMID: 21106008
-
The evolution of land plant cilia.New Phytol. 2012 Aug;195(3):526-540. doi: 10.1111/j.1469-8137.2012.04197.x. Epub 2012 Jun 12. New Phytol. 2012. PMID: 22691130 Review.
Cited by
-
Plastid phylogenomics and cytonuclear discordance in Rubioideae, Rubiaceae.PLoS One. 2024 May 20;19(5):e0302365. doi: 10.1371/journal.pone.0302365. eCollection 2024. PLoS One. 2024. PMID: 38768140 Free PMC article.
-
Silurian Climatic Zonation of Cryptospore, Trilete Spore and Plant Megafossils, with Emphasis on the Přídolí Epoch.Life (Basel). 2024 Feb 16;14(2):258. doi: 10.3390/life14020258. Life (Basel). 2024. PMID: 38398767 Free PMC article.
-
Mitochondrial RNA editing sites affect the phylogenetic reconstruction of gymnosperms.Plant Divers. 2023 Feb 23;45(4):485-489. doi: 10.1016/j.pld.2023.02.004. eCollection 2023 Jul. Plant Divers. 2023. PMID: 37601539 Free PMC article.
-
Characterizing conflict and congruence of molecular evolution across organellar genome sequences for phylogenetics in land plants.Front Plant Sci. 2023 Mar 30;14:1125107. doi: 10.3389/fpls.2023.1125107. eCollection 2023. Front Plant Sci. 2023. PMID: 37063179 Free PMC article.
-
Divergent evolutionary trajectories of bryophytes and tracheophytes from a complex common ancestor of land plants.Nat Ecol Evol. 2022 Nov;6(11):1634-1643. doi: 10.1038/s41559-022-01885-x. Epub 2022 Sep 29. Nat Ecol Evol. 2022. PMID: 36175544 Free PMC article.
References
-
- Bowman J.L. Walkabout on the long branches of plant evolution. Curr. Opin. Plant Biol. 2013;16:70–77. - PubMed
-
- Bowman J.L., Floyd S.K., Sakakibara K. Green genes - comparative genomics of the green branch of life. Cell. 2007;129:229–234. - PubMed
-
- Chang Y., Graham S.W. Inferring the higher-order phylogeny of mosses (Bryophyta) and relatives using a large, multigene plastid data set. Am. J. Botany. 2011;98:839–849. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources


