Network diffusion accurately models the relationship between structural and functional brain connectivity networks
- PMID: 24384152
- PMCID: PMC3951650
- DOI: 10.1016/j.neuroimage.2013.12.039
Network diffusion accurately models the relationship between structural and functional brain connectivity networks
Abstract
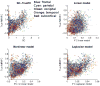
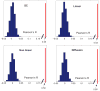
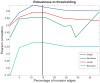
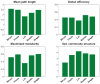
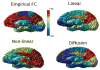
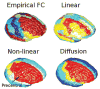
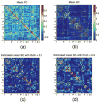
The relationship between anatomic connectivity of large-scale brain networks and their functional connectivity is of immense importance and an area of active research. Previous attempts have required complex simulations which model the dynamics of each cortical region, and explore the coupling between regions as derived by anatomic connections. While much insight is gained from these non-linear simulations, they can be computationally taxing tools for predicting functional from anatomic connectivities. Little attention has been paid to linear models. Here we show that a properly designed linear model appears to be superior to previous non-linear approaches in capturing the brain's long-range second order correlation structure that governs the relationship between anatomic and functional connectivities. We derive a linear network of brain dynamics based on graph diffusion, whereby the diffusing quantity undergoes a random walk on a graph. We test our model using subjects who underwent diffusion MRI and resting state fMRI. The network diffusion model applied to the structural networks largely predicts the correlation structures derived from their fMRI data, to a greater extent than other approaches. The utility of the proposed approach is that it can routinely be used to infer functional correlation from anatomic connectivity. And since it is linear, anatomic connectivity can also be inferred from functional data. The success of our model confirms the linearity of ensemble average signals in the brain, and implies that their long-range correlation structure may percolate within the brain via purely mechanistic processes enacted on its structural connectivity pathways.
Keywords: Brain connectivity; Functional connectivity; Networks; Structural connectivity.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures
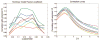


Similar articles
-
Changes in structural and functional connectivity among resting-state networks across the human lifespan.Neuroimage. 2014 Nov 15;102 Pt 2:345-57. doi: 10.1016/j.neuroimage.2014.07.067. Epub 2014 Aug 7. Neuroimage. 2014. PMID: 25109530
-
Functional brain connectivity is predictable from anatomic network's Laplacian eigen-structure.Neuroimage. 2018 May 15;172:728-739. doi: 10.1016/j.neuroimage.2018.02.016. Epub 2018 Feb 14. Neuroimage. 2018. PMID: 29454104 Free PMC article.
-
Linking human brain local activity fluctuations to structural and functional network architectures.Neuroimage. 2013 Jun;73:144-55. doi: 10.1016/j.neuroimage.2013.01.072. Epub 2013 Feb 8. Neuroimage. 2013. PMID: 23396160 Free PMC article.
-
Complex brain networks: graph theoretical analysis of structural and functional systems.Nat Rev Neurosci. 2009 Mar;10(3):186-98. doi: 10.1038/nrn2575. Epub 2009 Feb 4. Nat Rev Neurosci. 2009. PMID: 19190637 Review.
-
New approaches for exploring anatomical and functional connectivity in the human brain.Biol Psychiatry. 2004 Nov 1;56(9):613-9. doi: 10.1016/j.biopsych.2004.02.004. Biol Psychiatry. 2004. PMID: 15522243 Review.
Cited by
-
Adaptive rewiring: a general principle for neural network development.Front Netw Physiol. 2024 Oct 29;4:1410092. doi: 10.3389/fnetp.2024.1410092. eCollection 2024. Front Netw Physiol. 2024. PMID: 39534101 Free PMC article. Review.
-
The brain's first "traffic map" through Unified Structural and Functional Connectivity (USFC) modeling.Commun Biol. 2024 Nov 9;7(1):1477. doi: 10.1038/s42003-024-07160-y. Commun Biol. 2024. PMID: 39521849 Free PMC article.
-
Integrative, segregative, and degenerate harmonics of the structural connectome.Commun Biol. 2024 Aug 14;7(1):986. doi: 10.1038/s42003-024-06669-6. Commun Biol. 2024. PMID: 39143303 Free PMC article.
-
Simulation-based Inference of Developmental EEG Maturation with the Spectral Graph Model.ArXiv [Preprint]. 2024 Jul 26:arXiv:2405.02524v3. ArXiv. 2024. PMID: 39040639 Free PMC article. Preprint.
-
A multi-modal, asymmetric, weighted, and signed description of anatomical connectivity.Nat Commun. 2024 Jul 12;15(1):5865. doi: 10.1038/s41467-024-50248-6. Nat Commun. 2024. PMID: 38997282 Free PMC article.
References
-
- Agaskar A, Lu YM. An uncertainty principle for functions defined on graphs. Vol. 8138. SPIE; 2011. p. 81380T.
-
- Albright TD. Direction and orientation selectivity of neurons in visual area MT of the macaque. Journal of Neurophysiology. 1984;52:1106–1130. - PubMed
-
- Alemán-Gómez Y, Melie-García L, Valdés-Hernandez P. BASPM: Toolbox for automatic parcellation of brain structures. Human Brain Mapping 2006
-
- Almond D, Budd C, McCullen N. Emergent behaviour in large electrical networks. In: Georgoulis EH, Iske A, Levesley J, editors. Approximation Algorithms for Complex Systems, Vol. 3 of Springer Proceedings in Mathematics. Springer; Berlin Heidelberg: 2011. pp. 3–26.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources


