Isp7 is a novel regulator of amino acid uptake in the TOR signaling pathway
- PMID: 24344203
- PMCID: PMC4023818
- DOI: 10.1128/MCB.01473-13
Isp7 is a novel regulator of amino acid uptake in the TOR signaling pathway
Erratum in
- Mol Cell Biol. 2014 Apr;34(8):1535
Abstract
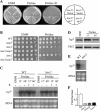
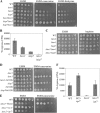
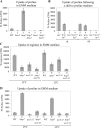
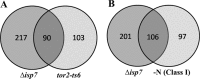
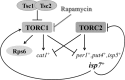
TOR proteins reside in two distinct complexes, TOR complexes 1 and 2 (TORC1 and TORC2), that are central for the regulation of cellular growth, proliferation, and survival. TOR is also the target for the immunosuppressive and anticancer drug rapamycin. In Schizosaccharomyces pombe, disruption of the TSC complex, mutations in which can lead to the tuberous sclerosis syndrome in humans, results in a rapamycin-sensitive phenotype under poor nitrogen conditions. We show here that the sensitivity to rapamycin is mediated via inhibition of TORC1 and suppressed by overexpression of isp7(+), a member of the family of 2-oxoglutarate-Fe(II)-dependent oxygenase genes. The transcript level of isp7(+) is negatively regulated by TORC1 but positively regulated by TORC2. Yet we find extensive similarity between the transcriptome of cells disrupted for isp7(+) and cells mutated in the catalytic subunit of TORC1. Moreover, Isp7 regulates amino acid permease expression in a fashion similar to that of TORC1 and opposite that of TORC2. Overexpression of isp7(+) induces TORC1-dependent phosphorylation of ribosomal protein Rps6 while inhibiting TORC2-dependent phosphorylation and activation of the AGC-like kinase Gad8. Taken together, our findings suggest a central role for Isp7 in amino acid homeostasis and the presence of isp7(+)-dependent regulatory loops that affect both TORC1 and TORC2.
Figures




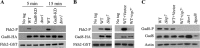
Similar articles
-
Fission Yeast TORC2 Signaling Pathway Ensures Cell Proliferation under Glucose-Limited, Nitrogen-Replete Conditions.Biomolecules. 2021 Oct 6;11(10):1465. doi: 10.3390/biom11101465. Biomolecules. 2021. PMID: 34680098 Free PMC article. Review.
-
Psk1, an AGC kinase family member in fission yeast, is directly phosphorylated and controlled by TORC1 and functions as S6 kinase.J Cell Sci. 2012 Dec 1;125(Pt 23):5840-9. doi: 10.1242/jcs.111146. Epub 2012 Sep 12. J Cell Sci. 2012. PMID: 22976295 Free PMC article.
-
Glucose activates TORC2-Gad8 protein via positive regulation of the cAMP/cAMP-dependent protein kinase A (PKA) pathway and negative regulation of the Pmk1 protein-mitogen-activated protein kinase pathway.J Biol Chem. 2014 Aug 1;289(31):21727-37. doi: 10.1074/jbc.M114.573824. Epub 2014 Jun 13. J Biol Chem. 2014. PMID: 24928510 Free PMC article.
-
TOR Complex 2- independent mutations in the regulatory PIF pocket of Gad8AKT1/SGK1 define separate branches of the stress response mechanisms in fission yeast.PLoS Genet. 2020 Nov 2;16(11):e1009196. doi: 10.1371/journal.pgen.1009196. eCollection 2020 Nov. PLoS Genet. 2020. PMID: 33137119 Free PMC article.
-
Express yourself: how PP2A-B55Pab1 helps TORC1 talk to TORC2.Curr Genet. 2018 Feb;64(1):43-51. doi: 10.1007/s00294-017-0721-8. Epub 2017 Jun 22. Curr Genet. 2018. PMID: 28643116 Free PMC article. Review.
Cited by
-
TOR regulates variability of protein synthesis rates.EMBO J. 2024 Apr;43(8):1618-1633. doi: 10.1038/s44318-024-00075-8. Epub 2024 Mar 18. EMBO J. 2024. PMID: 38499788 Free PMC article.
-
Interplays of AMPK and TOR in Autophagy Regulation in Yeast.Cells. 2023 Feb 4;12(4):519. doi: 10.3390/cells12040519. Cells. 2023. PMID: 36831186 Free PMC article. Review.
-
Cdc48 influence on separase levels is independent of mitosis and suggests translational sensitivity of separase.Cell Rep. 2022 Mar 22;38(12):110554. doi: 10.1016/j.celrep.2022.110554. Cell Rep. 2022. PMID: 35320724 Free PMC article.
-
Fission Yeast TORC2 Signaling Pathway Ensures Cell Proliferation under Glucose-Limited, Nitrogen-Replete Conditions.Biomolecules. 2021 Oct 6;11(10):1465. doi: 10.3390/biom11101465. Biomolecules. 2021. PMID: 34680098 Free PMC article. Review.
-
Genetic Analysis of Four Sexual Differentiation Process Proteins (isp4/SDPs) in Chaetomium thermophilum and Thermomyces lanuginosus Reveals Their Distinct Roles in Development.Front Microbiol. 2020 Jan 6;10:2994. doi: 10.3389/fmicb.2019.02994. eCollection 2019. Front Microbiol. 2020. PMID: 31969873 Free PMC article.
References
-
- Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. 2004. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 14:1296–1302. 10.1016/j.cub.2004.06.054 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

