Effects of diet on resource utilization by a model human gut microbiota containing Bacteroides cellulosilyticus WH2, a symbiont with an extensive glycobiome
- PMID: 23976882
- PMCID: PMC3747994
- DOI: 10.1371/journal.pbio.1001637
Effects of diet on resource utilization by a model human gut microbiota containing Bacteroides cellulosilyticus WH2, a symbiont with an extensive glycobiome
Abstract
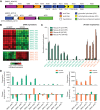
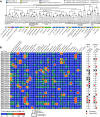
The human gut microbiota is an important metabolic organ, yet little is known about how its individual species interact, establish dominant positions, and respond to changes in environmental factors such as diet. In this study, gnotobiotic mice were colonized with an artificial microbiota comprising 12 sequenced human gut bacterial species and fed oscillating diets of disparate composition. Rapid, reproducible, and reversible changes in the structure of this assemblage were observed. Time-series microbial RNA-Seq analyses revealed staggered functional responses to diet shifts throughout the assemblage that were heavily focused on carbohydrate and amino acid metabolism. High-resolution shotgun metaproteomics confirmed many of these responses at a protein level. One member, Bacteroides cellulosilyticus WH2, proved exceptionally fit regardless of diet. Its genome encoded more carbohydrate active enzymes than any previously sequenced member of the Bacteroidetes. Transcriptional profiling indicated that B. cellulosilyticus WH2 is an adaptive forager that tailors its versatile carbohydrate utilization strategy to available dietary polysaccharides, with a strong emphasis on plant-derived xylans abundant in dietary staples like cereal grains. Two highly expressed, diet-specific polysaccharide utilization loci (PULs) in B. cellulosilyticus WH2 were identified, one with characteristics of xylan utilization systems. Introduction of a B. cellulosilyticus WH2 library comprising >90,000 isogenic transposon mutants into gnotobiotic mice, along with the other artificial community members, confirmed that these loci represent critical diet-specific fitness determinants. Carbohydrates that trigger dramatic increases in expression of these two loci and many of the organism's 111 other predicted PULs were identified by RNA-Seq during in vitro growth on 31 distinct carbohydrate substrates, allowing us to better interpret in vivo RNA-Seq and proteomics data. These results offer insight into how gut microbes adapt to dietary perturbations at both a community level and from the perspective of a well-adapted symbiont with exceptional saccharolytic capabilities, and illustrate the value of artificial communities.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Gnotobiotic mouse model of phage-bacterial host dynamics in the human gut.Proc Natl Acad Sci U S A. 2013 Dec 10;110(50):20236-41. doi: 10.1073/pnas.1319470110. Epub 2013 Nov 20. Proc Natl Acad Sci U S A. 2013. PMID: 24259713 Free PMC article.
-
Xylan degradation by the human gut Bacteroides xylanisolvens XB1A(T) involves two distinct gene clusters that are linked at the transcriptional level.BMC Genomics. 2016 May 4;17:326. doi: 10.1186/s12864-016-2680-8. BMC Genomics. 2016. PMID: 27142817 Free PMC article.
-
Bacteroides thetaiotaomicron metabolic activity decreases with polysaccharide molecular weight.mBio. 2024 Mar 13;15(3):e0259923. doi: 10.1128/mbio.02599-23. Epub 2024 Feb 20. mBio. 2024. PMID: 38376161 Free PMC article.
-
Navigating the Gut Buffet: Control of Polysaccharide Utilization in Bacteroides spp.Trends Microbiol. 2017 Dec;25(12):1005-1015. doi: 10.1016/j.tim.2017.06.009. Epub 2017 Jul 18. Trends Microbiol. 2017. PMID: 28733133 Review.
-
Glycan utilisation system in Bacteroides and Bifidobacteria and their roles in gut stability and health.Appl Microbiol Biotechnol. 2019 Sep;103(18):7287-7315. doi: 10.1007/s00253-019-10012-z. Epub 2019 Jul 22. Appl Microbiol Biotechnol. 2019. PMID: 31332487 Review.
Cited by
-
In vivo manipulation of human gut Bacteroides fitness by abiotic oligosaccharides.Nat Chem Biol. 2024 Oct 23. doi: 10.1038/s41589-024-01763-6. Online ahead of print. Nat Chem Biol. 2024. PMID: 39443715
-
Impact of ginsenoside Rb1 on gut microbiome and associated changes in pharmacokinetics in rats.Sci Rep. 2024 Sep 10;14(1):21168. doi: 10.1038/s41598-024-72225-1. Sci Rep. 2024. PMID: 39256599 Free PMC article.
-
The Utilization by Bacteroides spp. of a Purified Polysaccharide from Fuzhuan Brick Tea.Foods. 2024 May 26;13(11):1666. doi: 10.3390/foods13111666. Foods. 2024. PMID: 38890895 Free PMC article.
-
Polysaccharide utilization loci encoded DUF1735 likely functions as membrane-bound spacer for carbohydrate active enzymes.FEBS Open Bio. 2024 Jul;14(7):1133-1146. doi: 10.1002/2211-5463.13816. Epub 2024 May 12. FEBS Open Bio. 2024. PMID: 38735878 Free PMC article.
-
When simplicity triumphs: niche specialization of gut bacteria exists even for simple fiber structures.ISME Commun. 2024 Apr 11;4(1):ycae037. doi: 10.1093/ismeco/ycae037. eCollection 2024 Jan. ISME Commun. 2024. PMID: 38645272 Free PMC article.
References
-
- Flint HJ, Bayer EA, Rincon MT, Lamed R, White BA (2008) Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat Rev Microbiol 6: 121–131. - PubMed
-
- Roberfroid MB, Van Loo JA, Gibson GR (1998) The bifidogenic nature of chicory inulin and its hydrolysis products. J Nutr 128: 11–19. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases


