Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions
- PMID: 23598997
- PMCID: PMC3695513
- DOI: 10.1093/nar/gkt263
Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions
Abstract
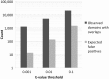
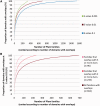
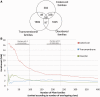
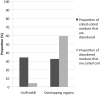
Detection of protein homology via sequence similarity has important applications in biology, from protein structure and function prediction to reconstruction of phylogenies. Although current methods for aligning protein sequences are powerful, challenges remain, including problems with homologous overextension of alignments and with regions under convergent evolution. Here, we test the ability of the profile hidden Markov model method HMMER3 to correctly assign homologous sequences to >13,000 manually curated families from the Pfam database. We identify problem families using protein regions that match two or more Pfam families not currently annotated as related in Pfam. We find that HMMER3 E-value estimates seem to be less accurate for families that feature periodic patterns of compositional bias, such as the ones typically observed in coiled-coils. These results support the continued use of manually curated inclusion thresholds in the Pfam database, especially on the subset of families that have been identified as problematic in experiments such as these. They also highlight the need for developing new methods that can correct for this particular type of compositional bias.
Figures
Similar articles
-
MACHOS: Markov clusters of homologous subsequences.Bioinformatics. 2008 Jul 1;24(13):i77-85. doi: 10.1093/bioinformatics/btn144. Bioinformatics. 2008. PMID: 18586748 Free PMC article.
-
Identifying protein domains with the Pfam database.Curr Protoc Bioinformatics. 2003 May;Chapter 2:Unit 2.5. doi: 10.1002/0471250953.bi0205s01. Curr Protoc Bioinformatics. 2003. PMID: 18428696
-
SUPFAM--a database of potential protein superfamily relationships derived by comparing sequence-based and structure-based families: implications for structural genomics and function annotation in genomes.Nucleic Acids Res. 2002 Jan 1;30(1):289-93. doi: 10.1093/nar/30.1.289. Nucleic Acids Res. 2002. PMID: 11752317 Free PMC article.
-
Sequence comparison and protein structure prediction.Curr Opin Struct Biol. 2006 Jun;16(3):374-84. doi: 10.1016/j.sbi.2006.05.006. Epub 2006 May 19. Curr Opin Struct Biol. 2006. PMID: 16713709 Review.
-
Pfam 10 years on: 10,000 families and still growing.Brief Bioinform. 2008 May;9(3):210-9. doi: 10.1093/bib/bbn010. Epub 2008 Mar 15. Brief Bioinform. 2008. PMID: 18344544 Review.
Cited by
-
Genomic insights into the cold adaptation and secondary metabolite potential of Pseudoalteromonas sp. WY3 from Antarctic krill.Front Microbiol. 2024 Nov 5;15:1459716. doi: 10.3389/fmicb.2024.1459716. eCollection 2024. Front Microbiol. 2024. PMID: 39564484 Free PMC article.
-
Expression of distal limb patterning genes in Hypsibius exemplaris indicate regionalization and suggest distal identity of tardigrade legs.Evodevo. 2024 Nov 13;15(1):15. doi: 10.1186/s13227-024-00235-1. Evodevo. 2024. PMID: 39538290 Free PMC article.
-
Structure-aware annotation of leucine-rich repeat domains.PLoS Comput Biol. 2024 Nov 5;20(11):e1012526. doi: 10.1371/journal.pcbi.1012526. eCollection 2024 Nov. PLoS Comput Biol. 2024. PMID: 39499733 Free PMC article.
-
Genome-wide analysis of the PYL gene family and identification of PYL genes that respond to cold stress in Triticum monococcum L. Subsp. Aegilopoides.Sci Rep. 2024 Nov 4;14(1):26627. doi: 10.1038/s41598-024-77962-x. Sci Rep. 2024. PMID: 39496812 Free PMC article.
-
Genome-wide analysis of WRKY gene family and the dynamic responses of key WRKY genes involved in cadmium stress in Brassica juncea.Front Plant Sci. 2024 Oct 10;15:1465905. doi: 10.3389/fpls.2024.1465905. eCollection 2024. Front Plant Sci. 2024. PMID: 39450073 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources


