Network-assisted investigation of combined causal signals from genome-wide association studies in schizophrenia
- PMID: 22792057
- PMCID: PMC3390381
- DOI: 10.1371/journal.pcbi.1002587
Network-assisted investigation of combined causal signals from genome-wide association studies in schizophrenia
Abstract
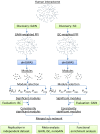
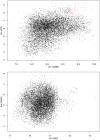
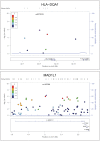
With the recent success of genome-wide association studies (GWAS), a wealth of association data has been accomplished for more than 200 complex diseases/traits, proposing a strong demand for data integration and interpretation. A combinatory analysis of multiple GWAS datasets, or an integrative analysis of GWAS data and other high-throughput data, has been particularly promising. In this study, we proposed an integrative analysis framework of multiple GWAS datasets by overlaying association signals onto the protein-protein interaction network, and demonstrated it using schizophrenia datasets. Building on a dense module search algorithm, we first searched for significantly enriched subnetworks for schizophrenia in each single GWAS dataset and then implemented a discovery-evaluation strategy to identify module genes with consistent association signals. We validated the module genes in an independent dataset, and also examined them through meta-analysis of the related SNPs using multiple GWAS datasets. As a result, we identified 205 module genes with a joint effect significantly associated with schizophrenia; these module genes included a number of well-studied candidate genes such as DISC1, GNA12, GNA13, GNAI1, GPR17, and GRIN2B. Further functional analysis suggested these genes are involved in neuronal related processes. Additionally, meta-analysis found that 18 SNPs in 9 module genes had P(meta)<1 × 10⁻⁴, including the gene HLA-DQA1 located in the MHC region on chromosome 6, which was reported in previous studies using the largest cohort of schizophrenia patients to date. These results demonstrated our bi-directional network-based strategy is efficient for identifying disease-associated genes with modest signals in GWAS datasets. This approach can be applied to any other complex diseases/traits where multiple GWAS datasets are available.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Searching joint association signals in CATIE schizophrenia genome-wide association studies through a refined integrative network approach.BMC Genomics. 2012;13 Suppl 6(Suppl 6):S15. doi: 10.1186/1471-2164-13-S6-S15. Epub 2012 Oct 26. BMC Genomics. 2012. PMID: 23134571 Free PMC article.
-
dmGWAS: dense module searching for genome-wide association studies in protein-protein interaction networks.Bioinformatics. 2011 Jan 1;27(1):95-102. doi: 10.1093/bioinformatics/btq615. Epub 2010 Nov 2. Bioinformatics. 2011. PMID: 21045073 Free PMC article.
-
Protein-interaction-network-based analysis for genome-wide association analysis of schizophrenia in Han Chinese population.J Psychiatr Res. 2014 Mar;50:73-8. doi: 10.1016/j.jpsychires.2013.11.014. Epub 2013 Dec 13. J Psychiatr Res. 2014. PMID: 24365204
-
Network.assisted analysis to prioritize GWAS results: principles, methods and perspectives.Hum Genet. 2014 Feb;133(2):125-38. doi: 10.1007/s00439-013-1377-1. Hum Genet. 2014. PMID: 24122152 Free PMC article. Review.
-
An integrative functional genomics approach for discovering biomarkers in schizophrenia.Brief Funct Genomics. 2011 Nov;10(6):387-99. doi: 10.1093/bfgp/elr036. Epub 2011 Dec 8. Brief Funct Genomics. 2011. PMID: 22155586 Free PMC article. Review.
Cited by
-
Nicotine Exposure in a Phencyclidine-Induced Mice Model of Schizophrenia: Sex-Selective Medial Prefrontal Cortex Protein Markers of the Combined Insults in Adolescent Mice.Int J Mol Sci. 2023 Sep 27;24(19):14634. doi: 10.3390/ijms241914634. Int J Mol Sci. 2023. PMID: 37834084 Free PMC article.
-
The genetic architecture of fornix white matter microstructure and their involvement in neuropsychiatric disorders.Transl Psychiatry. 2023 May 26;13(1):180. doi: 10.1038/s41398-023-02475-6. Transl Psychiatry. 2023. PMID: 37236919 Free PMC article.
-
scGWAS: landscape of trait-cell type associations by integrating single-cell transcriptomics-wide and genome-wide association studies.Genome Biol. 2022 Oct 17;23(1):220. doi: 10.1186/s13059-022-02785-w. Genome Biol. 2022. PMID: 36253801 Free PMC article.
-
Different responses to risperidone treatment in Schizophrenia: a multicenter genome-wide association and whole exome sequencing joint study.Transl Psychiatry. 2022 Apr 28;12(1):173. doi: 10.1038/s41398-022-01942-w. Transl Psychiatry. 2022. PMID: 35484098 Free PMC article.
-
Glial changes in schizophrenia: Genetic and epigenetic approach.Indian J Psychiatry. 2022 Jan-Feb;64(1):3-12. doi: 10.4103/indianjpsychiatry.indianjpsychiatry_104_21. Epub 2022 Jan 21. Indian J Psychiatry. 2022. PMID: 35400734 Free PMC article. Review.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous


