MYC on the path to cancer
- PMID: 22464321
- PMCID: PMC3345192
- DOI: 10.1016/j.cell.2012.03.003
MYC on the path to cancer
Abstract
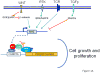
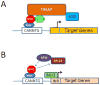
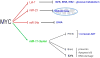
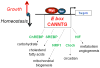
The MYC oncogene contributes to the genesis of many human cancers. Recent insights into its expression and function have led to therapeutic opportunities. MYC's activation by bromodomain proteins could be inhibited by drug-like molecules, resulting in tumor inhibition in vivo. Tumor growth can also be curbed by pharmacologically uncoupling bioenergetic pathways involving glucose or glutamine metabolism from Myc-induced cellular biomass accumulation. Other approaches to halt Myc on the path to cancer involve targeting Myc-Max dimerization or Myc-induced microRNA expression. Here the richness of our understanding of MYC is reviewed, highlighting new biological insights and opportunities for cancer therapies.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Drosophila Myc: A master regulator of cellular performance.Biochim Biophys Acta. 2015 May;1849(5):570-81. doi: 10.1016/j.bbagrm.2014.06.021. Epub 2014 Jul 8. Biochim Biophys Acta. 2015. PMID: 25010747 Free PMC article. Review.
-
The Molecular 'Myc-anisms' Behind Myc-Driven Tumorigenesis and the Relevant Myc-Directed Therapeutics.Int J Mol Sci. 2020 Dec 13;21(24):9486. doi: 10.3390/ijms21249486. Int J Mol Sci. 2020. PMID: 33322239 Free PMC article. Review.
-
Functional interactions among members of the MAX and MLX transcriptional network during oncogenesis.Biochim Biophys Acta. 2015 May;1849(5):484-500. doi: 10.1016/j.bbagrm.2014.05.016. Epub 2014 May 22. Biochim Biophys Acta. 2015. PMID: 24857747 Free PMC article. Review.
-
Myc's secret life without Max.Cell Cycle. 2009 Dec;8(23):3848-53. doi: 10.4161/cc.8.23.10088. Epub 2009 Dec 15. Cell Cycle. 2009. PMID: 19887915 Review.
-
MYC and transcription elongation.Cold Spring Harb Perspect Med. 2014 Jan 1;4(1):a020990. doi: 10.1101/cshperspect.a020990. Cold Spring Harb Perspect Med. 2014. PMID: 24384817 Free PMC article. Review.
Cited by
-
The 24-hour molecular landscape after exercise in humans reveals MYC is sufficient for muscle growth.EMBO Rep. 2024 Oct 31. doi: 10.1038/s44319-024-00299-z. Online ahead of print. EMBO Rep. 2024. PMID: 39482487
-
Global loss of promoter-enhancer connectivity and rebalancing of gene expression during early colorectal cancer carcinogenesis.Nat Cancer. 2024 Oct 30. doi: 10.1038/s43018-024-00823-z. Online ahead of print. Nat Cancer. 2024. PMID: 39478119
-
Transcriptional regulation by MYC: an emerging new model.Oncogene. 2024 Oct 28. doi: 10.1038/s41388-024-03174-2. Online ahead of print. Oncogene. 2024. PMID: 39468222 Review.
-
How lactate affects immune strategies in lymphoma.Front Mol Biosci. 2024 Oct 11;11:1480884. doi: 10.3389/fmolb.2024.1480884. eCollection 2024. Front Mol Biosci. 2024. PMID: 39464313 Free PMC article. Review.
-
Combinatorial Anti-Cancer Effect of Polypurine Reverse Hoogsteen Hairpins against KRAS and MYC Targeting in Prostate and Pancreatic Cancer Cell Lines.Genes (Basel). 2024 Oct 16;15(10):1332. doi: 10.3390/genes15101332. Genes (Basel). 2024. PMID: 39457457 Free PMC article.
References
-
- Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, Palmiter RD, Brinster RL. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–538. - PubMed
-
- Adhikary S, Marinoni F, Hock A, Hulleman E, Popov N, Beier R, Bernard S, Quarto M, Capra M, Goettig S, et al. The ubiquitin ligase HectH9 regulates transcriptional activation by Myc and is essential for tumor cell proliferation. Cell. 2005;123:409–421. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources


