Influenza research database: an integrated bioinformatics resource for influenza research and surveillance
- PMID: 22260278
- PMCID: PMC3345175
- DOI: 10.1111/j.1750-2659.2011.00331.x
Influenza research database: an integrated bioinformatics resource for influenza research and surveillance
Abstract
Background: The recent emergence of the 2009 pandemic influenza A/H1N1 virus has highlighted the value of free and open access to influenza virus genome sequence data integrated with information about other important virus characteristics.
Design: The Influenza Research Database (IRD, http://www.fludb.org) is a free, open, publicly-accessible resource funded by the U.S. National Institute of Allergy and Infectious Diseases through the Bioinformatics Resource Centers program. IRD provides a comprehensive, integrated database and analysis resource for influenza sequence, surveillance, and research data, including user-friendly interfaces for data retrieval, visualization and comparative genomics analysis, together with personal log in-protected 'workbench' spaces for saving data sets and analysis results. IRD integrates genomic, proteomic, immune epitope, and surveillance data from a variety of sources, including public databases, computational algorithms, external research groups, and the scientific literature.
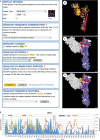
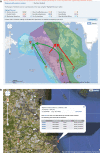
Results: To demonstrate the utility of the data and analysis tools available in IRD, two scientific use cases are presented. A comparison of hemagglutinin sequence conservation and epitope coverage information revealed highly conserved protein regions that can be recognized by the human adaptive immune system as possible targets for inducing cross-protective immunity. Phylogenetic and geospatial analysis of sequences from wild bird surveillance samples revealed a possible evolutionary connection between influenza virus from Delaware Bay shorebirds and Alberta ducks.
Conclusions: The IRD provides a wealth of integrated data and information about influenza virus to support research of the genetic determinants dictating virus pathogenicity, host range restriction and transmission, and to facilitate development of vaccines, diagnostics, and therapeutics.
© 2012 Blackwell Publishing Ltd.
Figures



Similar articles
-
Influenza Research Database: An integrated bioinformatics resource for influenza virus research.Nucleic Acids Res. 2017 Jan 4;45(D1):D466-D474. doi: 10.1093/nar/gkw857. Epub 2016 Sep 26. Nucleic Acids Res. 2017. PMID: 27679478 Free PMC article.
-
Public health. Surveillance of animal influenza for pandemic preparedness.Science. 2012 Mar 9;335(6073):1173-4. doi: 10.1126/science.1219936. Epub 2012 Feb 16. Science. 2012. PMID: 22345402 No abstract available.
-
The new influenza A H1N1 virus: balancing on the interface of humans and animals.Can Vet J. 2010 Jan;51(1):56-62. Can Vet J. 2010. PMID: 20357942 Free PMC article.
-
Swine flu--an epidemiological review.J Commun Dis. 2009 Sep;41(3):139-48. J Commun Dis. 2009. PMID: 22010480 Review. No abstract available.
-
[Epizootics of "avian" influenza as the manifestation of pandemic].Zh Mikrobiol Epidemiol Immunobiol. 2006 Jan-Feb;(1):81-7. Zh Mikrobiol Epidemiol Immunobiol. 2006. PMID: 16532652 Review. Russian.
Cited by
-
A systematic review of laboratory investigations into the pathogenesis of avian influenza viruses in wild avifauna of North America.Proc Biol Sci. 2024 Oct;291(2033):20241845. doi: 10.1098/rspb.2024.1845. Epub 2024 Oct 30. Proc Biol Sci. 2024. PMID: 39471857 Free PMC article.
-
Navigating the Landscape: A Comprehensive Review of Current Virus Databases.Viruses. 2023 Aug 29;15(9):1834. doi: 10.3390/v15091834. Viruses. 2023. PMID: 37766241 Free PMC article. Review.
-
Early Transcriptional Responses of Human Nasal Epithelial Cells to Infection with Influenza A and SARS-CoV-2 Virus Differ and Are Influenced by Physiological Temperature.Pathogens. 2023 Mar 18;12(3):480. doi: 10.3390/pathogens12030480. Pathogens. 2023. PMID: 36986402 Free PMC article.
-
H9N2 avian influenza virus dispersal along Bangladeshi poultry trading networks.Virus Evol. 2023 Feb 25;9(1):vead014. doi: 10.1093/ve/vead014. eCollection 2023. Virus Evol. 2023. PMID: 36968264 Free PMC article.
-
Early transcriptional responses of human nasal epithelial cells to infection with Influenza A and SARS-CoV-2 virus differ and are influenced by physiological temperature.bioRxiv [Preprint]. 2023 Mar 9:2023.03.07.531609. doi: 10.1101/2023.03.07.531609. bioRxiv. 2023. Update in: Pathogens. 2023 Mar 18;12(3):480. doi: 10.3390/pathogens12030480. PMID: 36945583 Free PMC article. Updated. Preprint.
References
-
- Palese P, Shaw ML. Orthomyxoviridae: the viruses and their replication; in Fields BN, Knipe DM, Howley PM. (eds): Fields’ Virology. Philadelphia: Wolters Kluwer Health/Lippincott Williams & Wilkins, 2007; 1647–1689.
-
- World Health Organization . Influenza web page. Available at http://www.who.int/mediacentre/factsheets/fs211/en/ (Accessed 22 July 2011).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical


