Representative proteomes: a stable, scalable and unbiased proteome set for sequence analysis and functional annotation
- PMID: 21556138
- PMCID: PMC3083393
- DOI: 10.1371/journal.pone.0018910
Representative proteomes: a stable, scalable and unbiased proteome set for sequence analysis and functional annotation
Abstract
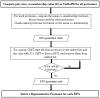
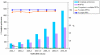
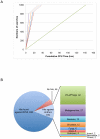
The accelerating growth in the number of protein sequences taxes both the computational and manual resources needed to analyze them. One approach to dealing with this problem is to minimize the number of proteins subjected to such analysis in a way that minimizes loss of information. To this end we have developed a set of Representative Proteomes (RPs), each selected from a Representative Proteome Group (RPG) containing similar proteomes calculated based on co-membership in UniRef50 clusters. A Representative Proteome is the proteome that can best represent all the proteomes in its group in terms of the majority of the sequence space and information. RPs at 75%, 55%, 35% and 15% co-membership threshold (CMT) are provided to allow users to decrease or increase the granularity of the sequence space based on their requirements. We find that a CMT of 55% (RP55) most closely follows standard taxonomic classifications. Further analysis of this set reveals that sequence space is reduced by more than 80% relative to UniProtKB, while retaining both sequence diversity (over 95% of InterPro domains) and annotation information (93% of experimentally characterized proteins). All sets can be browsed and are available for sequence similarity searches and download at http://www.proteininformationresource.org/rps, while the set of 637 RPs determined using a 55% CMT are also available for text searches. Potential applications include sequence similarity searches, protein classification and targeted protein annotation and characterization.
Conflict of interest statement
Figures

Similar articles
-
Computational clustering for viral reference proteomes.Bioinformatics. 2016 Jul 1;32(13):2041-3. doi: 10.1093/bioinformatics/btw110. Epub 2016 Feb 26. Bioinformatics. 2016. PMID: 27153712 Free PMC article.
-
Minimizing proteome redundancy in the UniProt Knowledgebase.Database (Oxford). 2016 Dec 26;2016:baw139. doi: 10.1093/database/baw139. Print 2016. Database (Oxford). 2016. PMID: 28025334 Free PMC article.
-
PCAS--a precomputed proteome annotation database resource.BMC Genomics. 2003 Nov 1;4(1):42. doi: 10.1186/1471-2164-4-42. BMC Genomics. 2003. PMID: 14594458 Free PMC article.
-
In silico characterization of proteins: UniProt, InterPro and Integr8.Mol Biotechnol. 2008 Feb;38(2):165-77. doi: 10.1007/s12033-007-9003-x. Epub 2007 Oct 4. Mol Biotechnol. 2008. PMID: 18219596 Review.
-
An Experimental Approach to Genome Annotation: This report is based on a colloquium sponsored by the American Academy of Microbiology held July 19-20, 2004, in Washington, DC.Washington (DC): American Society for Microbiology; 2004. Washington (DC): American Society for Microbiology; 2004. PMID: 33001599 Free Books & Documents. Review.
Cited by
-
Determinants of chemoselectivity in ubiquitination by the J2 family of ubiquitin-conjugating enzymes.EMBO J. 2024 Nov 12. doi: 10.1038/s44318-024-00301-3. Online ahead of print. EMBO J. 2024. PMID: 39533056
-
Structure-Informed Protein Language Models are Robust Predictors for Variant Effects.Res Sq [Preprint]. 2023 Aug 3:rs.3.rs-3219092. doi: 10.21203/rs.3.rs-3219092/v1. Res Sq. 2023. Update in: Hum Genet. 2024 Aug 8. doi: 10.1007/s00439-024-02695-w PMID: 37577664 Free PMC article. Updated. Preprint.
-
Developmental and temporal changes in petunia petal transcriptome reveal scent-repressing plant-specific RING-kinase-WD40 protein.Front Plant Sci. 2023 Jun 8;14:1180899. doi: 10.3389/fpls.2023.1180899. eCollection 2023. Front Plant Sci. 2023. PMID: 37360732 Free PMC article.
-
A general mechanism for transcription bubble nucleation in bacteria.Proc Natl Acad Sci U S A. 2023 Apr 4;120(14):e2220874120. doi: 10.1073/pnas.2220874120. Epub 2023 Mar 27. Proc Natl Acad Sci U S A. 2023. PMID: 36972428 Free PMC article.
-
On the Origin and Evolution of Microbial Mercury Methylation.Genome Biol Evol. 2023 Apr 6;15(4):evad051. doi: 10.1093/gbe/evad051. Genome Biol Evol. 2023. PMID: 36951100 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources


