Correlated z-values and the accuracy of large-scale statistical estimates
- PMID: 21052523
- PMCID: PMC2967047
- DOI: 10.1198/jasa.2010.tm09129
Correlated z-values and the accuracy of large-scale statistical estimates
Abstract
We consider large-scale studies in which there are hundreds or thousands of correlated cases to investigate, each represented by its own normal variate, typically a z-value. A familiar example is provided by a microarray experiment comparing healthy with sick subjects' expression levels for thousands of genes. This paper concerns the accuracy of summary statistics for the collection of normal variates, such as their empirical cdf or a false discovery rate statistic. It seems like we must estimate an N by N correlation matrix, N the number of cases, but our main result shows that this is not necessary: good accuracy approximations can be based on the root mean square correlation over all N · (N - 1)/2 pairs, a quantity often easily estimated. A second result shows that z-values closely follow normal distributions even under non-null conditions, supporting application of the main theorem. Practical application of the theory is illustrated for a large leukemia microarray study.
Figures
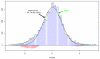

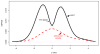
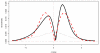
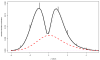

Similar articles
-
Selection of differentially expressed genes in microarray data analysis.Pharmacogenomics J. 2007 Jun;7(3):212-20. doi: 10.1038/sj.tpj.6500412. Epub 2006 Aug 29. Pharmacogenomics J. 2007. PMID: 16940966
-
Inference with Transposable Data: Modeling the Effects of Row and Column Correlations.J R Stat Soc Series B Stat Methodol. 2012 Sep;74(4):721-743. doi: 10.1111/j.1467-9868.2011.01027.x. Epub 2012 Mar 16. J R Stat Soc Series B Stat Methodol. 2012. PMID: 34880705 Free PMC article.
-
Estimation of false discovery rates in multiple testing: application to gene microarray data.Biometrics. 2003 Dec;59(4):1071-81. doi: 10.1111/j.0006-341x.2003.00123.x. Biometrics. 2003. PMID: 14969487
-
Applications of Monte Carlo Simulation in Modelling of Biochemical Processes.In: Mode CJ, editor. Applications of Monte Carlo Methods in Biology, Medicine and Other Fields of Science [Internet]. Rijeka (HR): InTech; 2011 Feb 28. Chapter 4. In: Mode CJ, editor. Applications of Monte Carlo Methods in Biology, Medicine and Other Fields of Science [Internet]. Rijeka (HR): InTech; 2011 Feb 28. Chapter 4. PMID: 28045483 Free Books & Documents. Review.
-
Rounding, but not randomization method, non-normality, or correlation, affected baseline P-value distributions in randomized trials.J Clin Epidemiol. 2019 Jun;110:50-62. doi: 10.1016/j.jclinepi.2019.03.001. Epub 2019 Mar 8. J Clin Epidemiol. 2019. PMID: 30858019 Review.
Cited by
-
UNIFYING AND GENERALIZING METHODS FOR REMOVING UNWANTED VARIATION BASED ON NEGATIVE CONTROLS.Stat Sin. 2021 Jul;31(3):1145-1166. doi: 10.5705/ss.202018.0345. Stat Sin. 2021. PMID: 38148787 Free PMC article.
-
Discussion of "Confidence Intervals for Nonparametric Empirical Bayes Analysis".J Am Stat Assoc. 2022;117(539):1186-1191. doi: 10.1080/01621459.2022.2093727. Epub 2022 Sep 12. J Am Stat Assoc. 2022. PMID: 37275677 Free PMC article. No abstract available.
-
A Test to Distinguish Monotone Homogeneity from Monotone Multifactor Models.Psychometrika. 2023 Jun;88(2):387-412. doi: 10.1007/s11336-023-09905-w. Epub 2023 Mar 18. Psychometrika. 2023. PMID: 36933110 Free PMC article.
-
Distinct sex-specific DNA methylation differences in Alzheimer's disease.Alzheimers Res Ther. 2022 Sep 15;14(1):133. doi: 10.1186/s13195-022-01070-z. Alzheimers Res Ther. 2022. PMID: 36109771 Free PMC article.
-
A resource for integrated genomic analysis of the human liver.Sci Rep. 2022 Sep 7;12(1):15151. doi: 10.1038/s41598-022-18506-z. Sci Rep. 2022. PMID: 36071064 Free PMC article.
References
-
- Bolstad B, Irizarry R, Astrand M, Speed T. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. - PubMed
-
- Clarke S, Hall P. Robustness of multiple testing procedures against dependence. Ann. Statist. 2009;37:332–358.
-
- Csörgő S, Mielniczuk J. The empirical process of a short-range dependent stationary sequence under Gaussian subordination. Probab. Theory Related Fields. 1996;104:15–25.
-
- Desai K, Deller J, McCormick J. The distribution of number of false discoveries for highly correlated null hypotheses. Ann. Appl. Statist. 2009 Submitted, under review.
-
- Dudoit S, Laan M. J. van der, Pollard KS. Multiple testing. I. Single-step procedures for control of general type I error rates. Stat. Appl. Genet. Mol. Biol. 2004;3:71. Art. 13. electronic. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

