Long noncoding RNA as modular scaffold of histone modification complexes
- PMID: 20616235
- PMCID: PMC2967777
- DOI: 10.1126/science.1192002
Long noncoding RNA as modular scaffold of histone modification complexes
Abstract
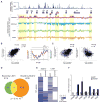
Long intergenic noncoding RNAs (lincRNAs) regulate chromatin states and epigenetic inheritance. Here, we show that the lincRNA HOTAIR serves as a scaffold for at least two distinct histone modification complexes. A 5' domain of HOTAIR binds polycomb repressive complex 2 (PRC2), whereas a 3' domain of HOTAIR binds the LSD1/CoREST/REST complex. The ability to tether two distinct complexes enables RNA-mediated assembly of PRC2 and LSD1 and coordinates targeting of PRC2 and LSD1 to chromatin for coupled histone H3 lysine 27 methylation and lysine 4 demethylation. Our results suggest that lincRNAs may serve as scaffolds by providing binding surfaces to assemble select histone modification enzymes, thereby specifying the pattern of histone modifications on target genes.
Figures



Similar articles
-
A model for transmission of the H3K27me3 epigenetic mark.Nat Cell Biol. 2008 Nov;10(11):1291-300. doi: 10.1038/ncb1787. Epub 2008 Oct 19. Nat Cell Biol. 2008. PMID: 18931660
-
Role of histone H3 lysine 27 methylation in Polycomb-group silencing.Science. 2002 Nov 1;298(5595):1039-43. doi: 10.1126/science.1076997. Epub 2002 Sep 26. Science. 2002. PMID: 12351676
-
Regulation of CD11b transcription by decreasing PRC2 and increased acH4 level during ATRA-induced HL-60 differentiation.Acta Biochim Biophys Sin (Shanghai). 2009 Jul;41(7):588-93. doi: 10.1093/abbs/gmp046. Acta Biochim Biophys Sin (Shanghai). 2009. PMID: 19578722
-
Inner workings and regulatory inputs that control Polycomb repressive complex 2.Chromosoma. 2012 Jun;121(3):221-34. doi: 10.1007/s00412-012-0361-1. Epub 2012 Feb 19. Chromosoma. 2012. PMID: 22349693 Free PMC article. Review.
-
Chromatin regulation: how complex does it get?Epigenetics. 2014 Nov;9(11):1485-95. doi: 10.4161/15592294.2014.971580. Epigenetics. 2014. PMID: 25482055 Free PMC article. Review.
Cited by
-
The Association and Prognostic Implications of Long Non-Coding RNAs in Major Psychiatric Disorders, Alzheimer's Diseases and Parkinson's Diseases: A Systematic Review.Int J Mol Sci. 2024 Oct 12;25(20):10995. doi: 10.3390/ijms252010995. Int J Mol Sci. 2024. PMID: 39456775 Free PMC article. Review.
-
SNHG6 facilitates the epithelial-mesenchymal transition and metastatic potential of esophageal squamous carcinoma through miR-26b-5p/ ITGB1 axis.Sci Rep. 2024 Oct 23;14(1):25005. doi: 10.1038/s41598-024-76521-8. Sci Rep. 2024. PMID: 39443675 Free PMC article.
-
siRNAs Targeting Non-Human Species-Specific lncRNAs Trigger Cell Death in Human Colorectal Cancer Cells.J Cancer. 2024 Sep 23;15(18):5956-5967. doi: 10.7150/jca.99462. eCollection 2024. J Cancer. 2024. PMID: 39440066 Free PMC article.
-
Advantages of the zebrafish tumor xenograft model: the evaluation of efficacy in cancer therapy and the application to the study of lncRNAs.Front Immunol. 2024 Sep 30;15:1483192. doi: 10.3389/fimmu.2024.1483192. eCollection 2024. Front Immunol. 2024. PMID: 39403375 Free PMC article. Review.
-
Long Non-Coding RNAs in Non-Alcoholic Fatty Liver Disease; Friends or Foes?Cell Biochem Biophys. 2024 Oct 8. doi: 10.1007/s12013-024-01555-8. Online ahead of print. Cell Biochem Biophys. 2024. PMID: 39377981 Review.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources


