GIGA: a simple, efficient algorithm for gene tree inference in the genomic age
- PMID: 20534164
- PMCID: PMC2905364
- DOI: 10.1186/1471-2105-11-312
GIGA: a simple, efficient algorithm for gene tree inference in the genomic age
Abstract
Background: Phylogenetic relationships between genes are not only of theoretical interest: they enable us to learn about human genes through the experimental work on their relatives in numerous model organisms from bacteria to fruit flies and mice. Yet the most commonly used computational algorithms for reconstructing gene trees can be inaccurate for numerous reasons, both algorithmic and biological. Additional information beyond gene sequence data has been shown to improve the accuracy of reconstructions, though at great computational cost.
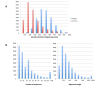
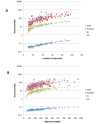
Results: We describe a simple, fast algorithm for inferring gene phylogenies, which makes use of information that was not available prior to the genomic age: namely, a reliable species tree spanning much of the tree of life, and knowledge of the complete complement of genes in a species' genome. The algorithm, called GIGA, constructs trees agglomeratively from a distance matrix representation of sequences, using simple rules to incorporate this genomic age information. GIGA makes use of a novel conceptualization of gene trees as being composed of orthologous subtrees (containing only speciation events), which are joined by other evolutionary events such as gene duplication or horizontal gene transfer. An important innovation in GIGA is that, at every step in the agglomeration process, the tree is interpreted/reinterpreted in terms of the evolutionary events that created it. Remarkably, GIGA performs well even when using a very simple distance metric (pairwise sequence differences) and no distance averaging over clades during the tree construction process.
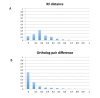
Conclusions: GIGA is efficient, allowing phylogenetic reconstruction of very large gene families and determination of orthologs on a large scale. It is exceptionally robust to adding more gene sequences, opening up the possibility of creating stable identifiers for referring to not only extant genes, but also their common ancestors. We compared trees produced by GIGA to those in the TreeFam database, and they were very similar in general, with most differences likely due to poor alignment quality. However, some remaining differences are algorithmic, and can be explained by the fact that GIGA tends to put a larger emphasis on minimizing gene duplication and deletion events.
Figures








Similar articles
-
On the quality of tree-based protein classification.Bioinformatics. 2005 May 1;21(9):1876-90. doi: 10.1093/bioinformatics/bti244. Epub 2005 Jan 12. Bioinformatics. 2005. PMID: 15647305
-
Algorithms for computing parsimonious evolutionary scenarios for genome evolution, the last universal common ancestor and dominance of horizontal gene transfer in the evolution of prokaryotes.BMC Evol Biol. 2003 Jan 6;3:2. doi: 10.1186/1471-2148-3-2. Epub 2003 Jan 6. BMC Evol Biol. 2003. PMID: 12515582 Free PMC article.
-
Genome trees constructed using five different approaches suggest new major bacterial clades.BMC Evol Biol. 2001 Oct 20;1:8. doi: 10.1186/1471-2148-1-8. BMC Evol Biol. 2001. PMID: 11734060 Free PMC article.
-
Genome trees and the tree of life.Trends Genet. 2002 Sep;18(9):472-9. doi: 10.1016/s0168-9525(02)02744-0. Trends Genet. 2002. PMID: 12175808 Review.
-
The inference of gene trees with species trees.Syst Biol. 2015 Jan;64(1):e42-62. doi: 10.1093/sysbio/syu048. Epub 2014 Jul 28. Syst Biol. 2015. PMID: 25070970 Free PMC article. Review.
Cited by
-
Stage-specific modulation of multinucleation, fusion, and resorption by the long non-coding RNA DLEU1 and miR-16 in human primary osteoclasts.Cell Death Dis. 2024 Oct 11;15(10):741. doi: 10.1038/s41419-024-06983-1. Cell Death Dis. 2024. PMID: 39389940 Free PMC article.
-
PANTHER: Making genome-scale phylogenetics accessible to all.Protein Sci. 2022 Jan;31(1):8-22. doi: 10.1002/pro.4218. Epub 2021 Nov 25. Protein Sci. 2022. PMID: 34717010 Free PMC article. Review.
-
Bayesian parameter estimation for automatic annotation of gene functions using observational data and phylogenetic trees.PLoS Comput Biol. 2021 Feb 18;17(2):e1007948. doi: 10.1371/journal.pcbi.1007948. eCollection 2021 Feb. PLoS Comput Biol. 2021. PMID: 33600408 Free PMC article.
-
PhyloGenes: An online phylogenetics and functional genomics resource for plant gene function inference.Plant Direct. 2020 Dec 30;4(12):e00293. doi: 10.1002/pld3.293. eCollection 2020 Dec. Plant Direct. 2020. PMID: 33392435 Free PMC article.
-
Unilateral L4-dorsal root ganglion stimulation evokes pain relief in chronic neuropathic postsurgical knee pain and changes of inflammatory markers: part II whole transcriptome profiling.J Transl Med. 2019 Jun 19;17(1):205. doi: 10.1186/s12967-019-1952-x. J Transl Med. 2019. PMID: 31217010 Free PMC article. Clinical Trial.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources


