Hidden dynamic allostery in a PDZ domain
- PMID: 19828436
- PMCID: PMC2775317
- DOI: 10.1073/pnas.0904492106
Hidden dynamic allostery in a PDZ domain
Abstract
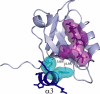
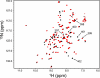
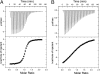
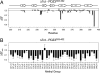
Structure-function relationships in proteins are predicated on the spatial proximity of noncovalently interacting groups of atoms. Thus, structural elements located away from a protein's active site are typically presumed to serve a stabilizing or scaffolding role for the larger structure. Here we report a functional role for a distal structural element in a PDZ domain, even though it is not required to maintain PDZ structure. The third PDZ domain from PSD-95/SAP90 (PDZ3) has an unusual additional third alpha helix (alpha3) that packs in contiguous fashion against the globular domain. Although alpha3 lies outside the active site and does not make direct contact with C-terminal peptide ligand, removal of alpha3 reduces ligand affinity by 21-fold. Further investigation revealed that the difference in binding free energies between the full-length and truncated constructs is predominantly entropic in nature and that without alpha3, picosecond-nanosecond side-chain dynamics are enhanced throughout the domain, as determined by (2)H methyl NMR relaxation. Thus, the distal modulation of binding function appears to occur via a delocalized conformational entropy mechanism. Without removal of alpha3 and characterization of side-chain dynamics, this dynamic allostery would have gone unnoticed. Moreover, what appeared at first to be an artificial modification of PDZ3 has been corroborated by experimentally verified phosphorylation of alpha3, revealing a tangible biological mechanism for this novel regulatory scheme. This hidden dynamic allostery raises the possibility of as-yet unidentified or untapped allosteric regulation in this PDZ domain and is a very clear example of function arising from dynamics rather than from structure.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Ensemble-Based Analysis of the Dynamic Allostery in the PSD-95 PDZ3 Domain in Relation to the General Variability of PDZ Structures.Int J Mol Sci. 2020 Nov 6;21(21):8348. doi: 10.3390/ijms21218348. Int J Mol Sci. 2020. PMID: 33172212 Free PMC article.
-
Modulation of the conformational landscape of the PDZ3 domain by perturbation on a distal non-canonical α3 helix: decoding the microscopic mechanism of allostery in the PDZ3 domain.Phys Chem Chem Phys. 2024 Aug 7;26(31):21249-21259. doi: 10.1039/d4cp01806k. Phys Chem Chem Phys. 2024. PMID: 39076021
-
The impact of extra-domain structures and post-translational modifications in the folding/misfolding behaviour of the third PDZ domain of MAGUK neuronal protein PSD-95.PLoS One. 2014 May 20;9(5):e98124. doi: 10.1371/journal.pone.0098124. eCollection 2014. PLoS One. 2014. PMID: 24845085 Free PMC article.
-
Allosterism in the PDZ Family.Int J Mol Sci. 2022 Jan 27;23(3):1454. doi: 10.3390/ijms23031454. Int J Mol Sci. 2022. PMID: 35163402 Free PMC article. Review.
-
Change in allosteric network affects binding affinities of PDZ domains: analysis through perturbation response scanning.PLoS Comput Biol. 2011 Oct;7(10):e1002154. doi: 10.1371/journal.pcbi.1002154. Epub 2011 Oct 6. PLoS Comput Biol. 2011. PMID: 21998559 Free PMC article. Review.
Cited by
-
Dynamic allostery drives autocrine and paracrine TGF-β signaling.Cell. 2024 Oct 31;187(22):6200-6219.e23. doi: 10.1016/j.cell.2024.08.036. Epub 2024 Sep 16. Cell. 2024. PMID: 39288764 Free PMC article.
-
Mapping protein conformational landscapes from crystallographic drug fragment screens.bioRxiv [Preprint]. 2024 Jul 30:2024.07.29.605395. doi: 10.1101/2024.07.29.605395. bioRxiv. 2024. PMID: 39131376 Free PMC article. Preprint.
-
Rapid Protein-Ligand Affinity Determination by Photoinduced Hyperpolarized NMR.J Am Chem Soc. 2024 Jul 3;146(26):17974-17985. doi: 10.1021/jacs.4c04000. Epub 2024 Jun 18. J Am Chem Soc. 2024. PMID: 38957136 Free PMC article.
-
Targeting TrkB-PSD-95 coupling to mitigate neurological disorders.Neural Regen Res. 2025 Mar 1;20(3):715-724. doi: 10.4103/NRR.NRR-D-23-02000. Epub 2024 May 13. Neural Regen Res. 2025. PMID: 38886937 Free PMC article.
-
Informational Analysis and Prediction of Obsessive-Compulsive Disorder Pathogenesis.Psychiatry Investig. 2024 May;21(5):464-474. doi: 10.30773/pi.2023.0149. Epub 2024 May 23. Psychiatry Investig. 2024. PMID: 38810995 Free PMC article.
References
-
- Zhang MJ, Wang WN. Organization of signaling complexes by PDZ-domain scaffold proteins. Acc Chem Res. 2003;36:530–538. - PubMed
-
- Garner CC, Nash J, Huganir RL. PDZ domains in synapse assembly and signalling. Trends Cell Biol. 2000;10:274–280. - PubMed
-
- Kim E, Sheng M. PDZ domain proteins of synapses. Nat Rev Neurosci. 2004;5:771–781. - PubMed
-
- Doyle DA, et al. Crystal structures of a complexed and peptide-free membrane protein-binding domain: Molecular basis of peptide recognition by PDZ. Cell. 1996;85:1067–1076. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases


