The salivary gland transcriptome of the neotropical malaria vector Anopheles darlingi reveals accelerated evolution of genes relevant to hematophagy
- PMID: 19178717
- PMCID: PMC2644710
- DOI: 10.1186/1471-2164-10-57
The salivary gland transcriptome of the neotropical malaria vector Anopheles darlingi reveals accelerated evolution of genes relevant to hematophagy
Abstract
Background: Mosquito saliva, consisting of a mixture of dozens of proteins affecting vertebrate hemostasis and having sugar digestive and antimicrobial properties, helps both blood and sugar meal feeding. Culicine and anopheline mosquitoes diverged ~150 MYA, and within the anophelines, the New World species diverged from those of the Old World ~95 MYA. While the sialotranscriptome (from the Greek sialo, saliva) of several species of the Cellia subgenus of Anopheles has been described thoroughly, no detailed analysis of any New World anopheline has been done to date. Here we present and analyze data from a comprehensive salivary gland (SG) transcriptome of the neotropical malaria vector Anopheles darlingi (subgenus Nyssorhynchus).
Results: A total of 2,371 clones randomly selected from an adult female An. darlingi SG cDNA library were sequenced and used to assemble a database that yielded 966 clusters of related sequences, 739 of which were singletons. Primer extension experiments were performed in selected clones to further extend sequence coverage, allowing for the identification of 183 protein sequences, 114 of which code for putative secreted proteins.
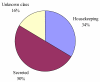
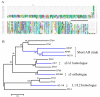
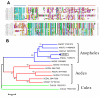
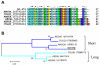
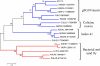
Conclusion: Comparative analysis of sialotranscriptomes of An. darlingi and An. gambiae reveals significant divergence of salivary proteins. On average, salivary proteins are only 53% identical, while housekeeping proteins are 86% identical between the two species. Furthermore, An. darlingi proteins were found that match culicine but not anopheline proteins, indicating loss or rapid evolution of these proteins in the old world Cellia subgenus. On the other hand, several well represented salivary protein families in old world anophelines are not expressed in An. darlingi.
Figures

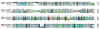

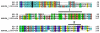
Similar articles
-
An insight into the sialotranscriptome of the West Nile mosquito vector, Culex tarsalis.BMC Genomics. 2010 Jan 20;11:51. doi: 10.1186/1471-2164-11-51. BMC Genomics. 2010. PMID: 20089177 Free PMC article.
-
An insight into the sialome of Anopheles funestus reveals an emerging pattern in anopheline salivary protein families.Insect Biochem Mol Biol. 2007 Feb;37(2):164-75. doi: 10.1016/j.ibmb.2006.11.005. Epub 2006 Nov 22. Insect Biochem Mol Biol. 2007. PMID: 17244545 Free PMC article.
-
Anopheline salivary protein genes and gene families: an evolutionary overview after the whole genome sequence of sixteen Anopheles species.BMC Genomics. 2017 Feb 13;18(1):153. doi: 10.1186/s12864-017-3579-8. BMC Genomics. 2017. PMID: 28193177 Free PMC article.
-
Salivary gland-specific gene expression in the malaria vector Anopheles gambiae.Parassitologia. 1999 Sep;41(1-3):483-7. Parassitologia. 1999. PMID: 10697906 Review.
-
Positive selection in multiple salivary gland proteins of Anophelinae reveals potential targets for vector control.Infect Genet Evol. 2022 Jun;100:105271. doi: 10.1016/j.meegid.2022.105271. Epub 2022 Mar 23. Infect Genet Evol. 2022. PMID: 35339698 Review.
Cited by
-
An insight into the female and male Sabethes cyaneus mosquito salivary glands transcriptome.Insect Biochem Mol Biol. 2023 Feb;153:103898. doi: 10.1016/j.ibmb.2022.103898. Epub 2022 Dec 30. Insect Biochem Mol Biol. 2023. PMID: 36587808 Free PMC article.
-
Novel salivary antihemostatic activities of long-form D7 proteins from the malaria vector Anopheles gambiae facilitate hematophagy.J Biol Chem. 2022 Jun;298(6):101971. doi: 10.1016/j.jbc.2022.101971. Epub 2022 Apr 20. J Biol Chem. 2022. PMID: 35460690 Free PMC article.
-
First evaluation of antibody responses to Culex quinquefasciatus salivary antigens as a serological biomarker of human exposure to Culex bites: A pilot study in Côte d'Ivoire.PLoS Negl Trop Dis. 2021 Dec 13;15(12):e0010004. doi: 10.1371/journal.pntd.0010004. eCollection 2021 Dec. PLoS Negl Trop Dis. 2021. PMID: 34898609 Free PMC article.
-
MicroRNAs from saliva of anopheline mosquitoes mimic human endogenous miRNAs and may contribute to vector-host-pathogen interactions.Sci Rep. 2019 Feb 27;9(1):2955. doi: 10.1038/s41598-019-39880-1. Sci Rep. 2019. PMID: 30814633 Free PMC article.
-
Immunization with AgTRIO, a Protein in Anopheles Saliva, Contributes to Protection against Plasmodium Infection in Mice.Cell Host Microbe. 2018 Apr 11;23(4):523-535.e5. doi: 10.1016/j.chom.2018.03.008. Cell Host Microbe. 2018. PMID: 29649443 Free PMC article.
References
-
- Ribeiro JMC. Blood-feeding arthropods: Live syringes or invertebrate pharmacologists? Infect Agents Dis. 1995;4:143–152. - PubMed
-
- Marinotti O, James AA. An alpha-glucosidase in the salivary glands of the vector mosquito, Aedes aegypti. Insect Biochem. 1990;20:619–623. doi: 10.1016/0020-1790(90)90074-5. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources


