High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor
- PMID: 17962520
- PMCID: PMC2583103
- DOI: 10.1126/science.1150577
High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor
Abstract
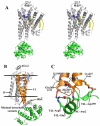
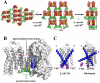
Heterotrimeric guanine nucleotide-binding protein (G protein)-coupled receptors constitute the largest family of eukaryotic signal transduction proteins that communicate across the membrane. We report the crystal structure of a human beta2-adrenergic receptor-T4 lysozyme fusion protein bound to the partial inverse agonist carazolol at 2.4 angstrom resolution. The structure provides a high-resolution view of a human G protein-coupled receptor bound to a diffusible ligand. Ligand-binding site accessibility is enabled by the second extracellular loop, which is held out of the binding cavity by a pair of closely spaced disulfide bridges and a short helical segment within the loop. Cholesterol, a necessary component for crystallization, mediates an intriguing parallel association of receptor molecules in the crystal lattice. Although the location of carazolol in the beta2-adrenergic receptor is very similar to that of retinal in rhodopsin, structural differences in the ligand-binding site and other regions highlight the challenges in using rhodopsin as a template model for this large receptor family.
Figures




Comment in
-
Biochemistry. Signaling across the cell membrane.Science. 2007 Nov 23;318(5854):1253-4. doi: 10.1126/science.1151656. Science. 2007. PMID: 18033872 No abstract available.
Similar articles
-
GPCR engineering yields high-resolution structural insights into beta2-adrenergic receptor function.Science. 2007 Nov 23;318(5854):1266-73. doi: 10.1126/science.1150609. Epub 2007 Oct 25. Science. 2007. PMID: 17962519
-
Biochemistry. Signaling across the cell membrane.Science. 2007 Nov 23;318(5854):1253-4. doi: 10.1126/science.1151656. Science. 2007. PMID: 18033872 No abstract available.
-
Crystallization of G protein-coupled receptors.Methods Cell Biol. 2013;117:451-68. doi: 10.1016/B978-0-12-408143-7.00024-4. Methods Cell Biol. 2013. PMID: 24143992 Free PMC article.
-
Structural features of β2 adrenergic receptor: crystal structures and beyond.Mol Cells. 2015;38(2):105-11. doi: 10.14348/molcells.2015.2301. Epub 2014 Dec 24. Mol Cells. 2015. PMID: 25537861 Free PMC article. Review.
-
G protein-coupled receptors--recent advances.Acta Biochim Pol. 2012;59(4):515-29. Epub 2012 Dec 18. Acta Biochim Pol. 2012. PMID: 23251911 Free PMC article. Review.
Cited by
-
VPS26 Moonlights as a β-Arrestin-like Adapter for a 7-Transmembrane RGS Protein in Arabidopsis thaliana.Biochemistry. 2024 Nov 19;63(22):2990-2999. doi: 10.1021/acs.biochem.4c00361. Epub 2024 Oct 28. Biochemistry. 2024. PMID: 39467170 Free PMC article.
-
Active state structures of a bistable visual opsin bound to G proteins.Nat Commun. 2024 Oct 16;15(1):8928. doi: 10.1038/s41467-024-53208-2. Nat Commun. 2024. PMID: 39414813 Free PMC article.
-
Integrative residue-intuitive machine learning and MD Approach to Unveil Allosteric Site and Mechanism for β2AR.Nat Commun. 2024 Sep 16;15(1):8130. doi: 10.1038/s41467-024-52399-y. Nat Commun. 2024. PMID: 39285201 Free PMC article.
-
Interaction and dynamics of chemokine receptor CXCR4 binding with CXCL12 and hBD-3.Commun Chem. 2024 Sep 13;7(1):205. doi: 10.1038/s42004-024-01280-6. Commun Chem. 2024. PMID: 39271963 Free PMC article.
-
Early Events in β2AR Dimer Dynamics Mediated by Activation-Related Microswitches.J Membr Biol. 2024 Dec;257(5-6):323-344. doi: 10.1007/s00232-024-00324-1. Epub 2024 Sep 6. J Membr Biol. 2024. PMID: 39240374
References
-
- Takeda S, Kadowaki S, Haga T, Takaesu H, Mitaku S. FEBS Lett. 2002;520:97. - PubMed
-
- Fredriksson R, Lagerstrom MC, Lundin LG, Schioth HB. Mol Pharmacol. 2003;63:1256. - PubMed
-
- Pierce KL, Premont RT, Lefkowitz RJ. Nat Rev Mol Cell Biol. 2002;3:639. - PubMed
-
- Lefkowitz RJ, Shenoy SK. Science. 2005;308:512. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- Y1-GM-1104/GM/NIGMS NIH HHS/United States
- F32 GM082028/GM/NIGMS NIH HHS/United States
- P50 GM073197/GM/NIGMS NIH HHS/United States
- P50 GM062411/GM/NIGMS NIH HHS/United States
- R21 GM075811/GM/NIGMS NIH HHS/United States
- GM075915/GM/NIGMS NIH HHS/United States
- R01 NS028471/NS/NINDS NIH HHS/United States
- U54 GM074961/GM/NIGMS NIH HHS/United States
- NS028471/NS/NINDS NIH HHS/United States
- U54 GM074961-030001/GM/NIGMS NIH HHS/United States
- R01 GM089857/GM/NIGMS NIH HHS/United States
- Y1-CO-1020/CO/NCI NIH HHS/United States
- P50 GM073197-04/GM/NIGMS NIH HHS/United States
- R01 GM056169/GM/NIGMS NIH HHS/United States
- R37 NS028471/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases


