Transcriptional regulation of the CO2-concentrating mechanism in a euryhaline, coastal marine cyanobacterium, Synechococcus sp. Strain PCC 7002: role of NdhR/CcmR
- PMID: 17307862
- PMCID: PMC1855907
- DOI: 10.1128/JB.01745-06
Transcriptional regulation of the CO2-concentrating mechanism in a euryhaline, coastal marine cyanobacterium, Synechococcus sp. Strain PCC 7002: role of NdhR/CcmR
Abstract
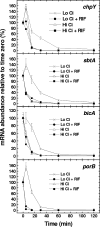
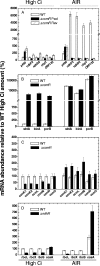
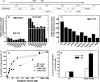
Cyanobacterial photosynthesis occurs in radically diverse habitats and utilizes various forms of a CO(2)-concentrating mechanism (CCM) featuring multiple inorganic carbon (C(i)) transporters. Cyanobacteria from dynamic environments can transform CCM activity depending on C(i) availability, and yet the molecular basis for this regulation is unclear, especially in coastal strains. LysR family transcription factors resembling the Calvin cycle regulator CbbR from proteobacteria have been implicated in the expression of C(i) transporter genes in freshwater cyanobacteria. Our survey of related factors revealed a group of divergent CbbR-like sequences confined to freshwater and coastal or offshore cyanobacteria. Inactivation of the single gene (termed ccmR) from this variable cluster in the euryhaline (coastal) strain Synechococcus sp. strain PCC 7002 led to constitutive expression of a high-affinity CCM. Derepression of HCO(3)(-) transporter gene transcription, including that of BicA, a recently discovered HCO(3)(-) transporter (G. D. Price et al., Proc. Natl. Acad. Sci. USA 101:18228-18233, 2004), was observed. A unique CcmR-regulated operon containing bicA plus 9 open reading frames encoding likely Na(+)/H(+) antiporters from the CPA1 and Mnh families was defined that is essential for maximal HCO(3)(-)-dependent oxygen evolution. The promoter region required for C(i)-regulated transcription of this operon was defined. We propose that CcmR (and its associated regulon) represents a specialization for species inhabiting environments subject to fluctuating C(i) concentrations.
Figures



Similar articles
-
Involvement of a CbbR homolog in low CO2-induced activation of the bicarbonate transporter operon in cyanobacteria.J Bacteriol. 2001 Mar;183(6):1891-8. doi: 10.1128/JB.183.6.1891-1898.2001. J Bacteriol. 2001. PMID: 11222586 Free PMC article.
-
Consequences of ccmR deletion on respiration, fermentation and H2 metabolism in cyanobacterium Synechococcus sp. PCC 7002.Biotechnol Bioeng. 2016 Jul;113(7):1448-59. doi: 10.1002/bit.25913. Epub 2016 Jan 28. Biotechnol Bioeng. 2016. PMID: 26704377
-
The CO2-concentrating mechanism of Synechococcus WH5701 is composed of native and horizontally-acquired components.Photosynth Res. 2011 Sep;109(1-3):59-72. doi: 10.1007/s11120-011-9641-5. Epub 2011 Mar 8. Photosynth Res. 2011. PMID: 21384181
-
Inorganic carbon transporters of the cyanobacterial CO2 concentrating mechanism.Photosynth Res. 2011 Sep;109(1-3):47-57. doi: 10.1007/s11120-010-9608-y. Epub 2011 Feb 26. Photosynth Res. 2011. PMID: 21359551 Review.
-
Genetics and control of CO(2) assimilation in the chemoautotroph Ralstonia eutropha.Arch Microbiol. 2002 Aug;178(2):85-93. doi: 10.1007/s00203-002-0441-3. Epub 2002 Jun 14. Arch Microbiol. 2002. PMID: 12115053 Review.
Cited by
-
Machine learning reveals the transcriptional regulatory network and circadian dynamics of Synechococcus elongatus PCC 7942.Proc Natl Acad Sci U S A. 2024 Sep 17;121(38):e2410492121. doi: 10.1073/pnas.2410492121. Epub 2024 Sep 13. Proc Natl Acad Sci U S A. 2024. PMID: 39269777 Free PMC article.
-
CupAR negatively controls the key protein CupA in the carbon acquisition complex NDH-1MS in Synechocystis sp. PCC 6803.J Biol Chem. 2024 Sep;300(9):107716. doi: 10.1016/j.jbc.2024.107716. Epub 2024 Aug 22. J Biol Chem. 2024. PMID: 39181331 Free PMC article.
-
Adapting from Low to High: An Update to CO2-Concentrating Mechanisms of Cyanobacteria and Microalgae.Plants (Basel). 2023 Apr 6;12(7):1569. doi: 10.3390/plants12071569. Plants (Basel). 2023. PMID: 37050194 Free PMC article. Review.
-
Stress Signaling in Cyanobacteria: A Mechanistic Overview.Life (Basel). 2020 Nov 26;10(12):312. doi: 10.3390/life10120312. Life (Basel). 2020. PMID: 33256109 Free PMC article. Review.
-
Overexpression of bicarbonate transporters in the marine cyanobacterium Synechococcus sp. PCC 7002 increases growth rate and glycogen accumulation.Biotechnol Biofuels. 2020 Jan 28;13:17. doi: 10.1186/s13068-020-1656-8. eCollection 2020. Biotechnol Biofuels. 2020. PMID: 32015756 Free PMC article.
References
-
- Andersson, C. R., N. F. Tsinoremas, J. Shelton, N. V. Lebedeva, J. Yarrow, H. T. Min, and S. S. Golden. 2000. Application of bioluminescence to the study of circadian rhythms in cyanobacteria. Methods Enzymol. 305:527-542. - PubMed
-
- Badger, M. R., K. Palmqvist, and J. W. Yu. 1994. Measurement of CO2 and HCO3− fluxes in cyanobacteria and microalgae during steady-state photosynthesis. Physiol. Plant 90:529-536.
-
- Badger, M. R., and G. D. Price. 2003. CO2 concentrating mechanisms in cyanobacteria: molecular components, their diversity and evolution. J. Exp. Bot. 54:609-622. - PubMed
-
- Badger, M. R., G. D. Price, B. M. Long, and F. J. Woodger. 2006. The environmental plasticity and ecological genomics of the cyanobacterial CO2 concentrating mechanism. J. Exp. Bot. 57:249-265. - PubMed
-
- Blanco-Rivero, A., F. Leganés, E. Fernández-Valiente, P. Calle, and F. Fernádez-Piñas. 2005. mrpA, a gene with roles in resistance to Na+ and adaptation to alkaline pH in the cyanobacterium Anabaena sp. PCC 7120. Microbiol. 151:1671-1682. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases


