Expansion of the BioCyc collection of pathway/genome databases to 160 genomes
- PMID: 16246909
- PMCID: PMC1266070
- DOI: 10.1093/nar/gki892
Expansion of the BioCyc collection of pathway/genome databases to 160 genomes
Abstract
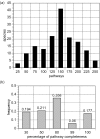
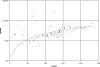
The BioCyc database collection is a set of 160 pathway/genome databases (PGDBs) for most eukaryotic and prokaryotic species whose genomes have been completely sequenced to date. Each PGDB in the BioCyc collection describes the genome and predicted metabolic network of a single organism, inferred from the MetaCyc database, which is a reference source on metabolic pathways from multiple organisms. In addition, each bacterial PGDB includes predicted operons for the corresponding species. The BioCyc collection provides a unique resource for computational systems biology, namely global and comparative analyses of genomes and metabolic networks, and a supplement to the BioCyc resource of curated PGDBs. The Omics viewer available through the BioCyc website allows scientists to visualize combinations of gene expression, proteomics and metabolomics data on the metabolic maps of these organisms. This paper discusses the computational methodology by which the BioCyc collection has been expanded, and presents an aggregate analysis of the collection that includes the range of number of pathways present in these organisms, and the most frequently observed pathways. We seek scientists to adopt and curate individual PGDBs within the BioCyc collection. Only by harnessing the expertise of many scientists we can hope to produce biological databases, which accurately reflect the depth and breadth of knowledge that the biomedical research community is producing.
Figures
Similar articles
-
The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases.Nucleic Acids Res. 2010 Jan;38(Database issue):D473-9. doi: 10.1093/nar/gkp875. Epub 2009 Oct 22. Nucleic Acids Res. 2010. PMID: 19850718 Free PMC article.
-
The MetaCyc Database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases.Nucleic Acids Res. 2008 Jan;36(Database issue):D623-31. doi: 10.1093/nar/gkm900. Epub 2007 Oct 27. Nucleic Acids Res. 2008. PMID: 17965431 Free PMC article.
-
The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases.Nucleic Acids Res. 2012 Jan;40(Database issue):D742-53. doi: 10.1093/nar/gkr1014. Epub 2011 Nov 18. Nucleic Acids Res. 2012. PMID: 22102576 Free PMC article.
-
An Experimental Approach to Genome Annotation: This report is based on a colloquium sponsored by the American Academy of Microbiology held July 19-20, 2004, in Washington, DC.Washington (DC): American Society for Microbiology; 2004. Washington (DC): American Society for Microbiology; 2004. PMID: 33001599 Free Books & Documents. Review.
-
DOOR: a prokaryotic operon database for genome analyses and functional inference.Brief Bioinform. 2019 Jul 19;20(4):1568-1577. doi: 10.1093/bib/bbx088. Brief Bioinform. 2019. PMID: 28968679 Review.
Cited by
-
Metalloproteomics Reveals Multi-Level Stress Response in Escherichia coli When Exposed to Arsenite.Int J Mol Sci. 2024 Sep 2;25(17):9528. doi: 10.3390/ijms25179528. Int J Mol Sci. 2024. PMID: 39273475 Free PMC article.
-
Patterns of change in regulatory modules of chemical reaction systems induced by network modification.PNAS Nexus. 2023 Dec 29;3(1):pgad441. doi: 10.1093/pnasnexus/pgad441. eCollection 2024 Jan. PNAS Nexus. 2023. PMID: 38292559 Free PMC article.
-
Multi-level advances in databases related to systems pharmacology in traditional Chinese medicine: a 60-year review.Front Pharmacol. 2023 Nov 14;14:1289901. doi: 10.3389/fphar.2023.1289901. eCollection 2023. Front Pharmacol. 2023. PMID: 38035021 Free PMC article. Review.
-
A Protocol for the Automatic Construction of Highly Curated Genome-Scale Models of Human Metabolism.Bioengineering (Basel). 2023 May 10;10(5):576. doi: 10.3390/bioengineering10050576. Bioengineering (Basel). 2023. PMID: 37237646 Free PMC article.
-
The inside scoop: Comparative genomics of two intranuclear bacteria, "Candidatus Berkiella cookevillensis" and "Candidatus Berkiella aquae".PLoS One. 2022 Dec 30;17(12):e0278206. doi: 10.1371/journal.pone.0278206. eCollection 2022. PLoS One. 2022. PMID: 36584052 Free PMC article.
References
-
- Romero P., Karp P. PseudoCyc, a pathway-genome database for Pseudomonas aeruginosa. J. Mol. Microbiol. Biotechnol. 2003;5:230–239. - PubMed
-
- Christie K.R., Weng S., Balakrishnan R., Costanzo M.C., Dolinski K., Dwight S.S., Engel S.R., Feierbach B., Fisk D.G., Hirschman J.E., et al. Saccharomyces Genome Database (SGD) provides tools to identify and analyze sequences from Saccharomyces cerevisiae and related sequences from other organisms. Nucleic Acids Res. 2004;32:D311–D314. - PMC - PubMed
-
- Rhee S.Y., Beavis W., Berardini T.Z., Chen G., Dixon D., Doyle A., Garcia-Hernandez M., Huala E., Lander G., Montoya M., et al. The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res. 2003;31:224–228. - PMC - PubMed


