Nanoarchaea: representatives of a novel archaeal phylum or a fast-evolving euryarchaeal lineage related to Thermococcales?
- PMID: 15892870
- PMCID: PMC1175954
- DOI: 10.1186/gb-2005-6-5-r42
Nanoarchaea: representatives of a novel archaeal phylum or a fast-evolving euryarchaeal lineage related to Thermococcales?
Abstract
Background: Cultivable archaeal species are assigned to two phyla -- the Crenarchaeota and the Euryarchaeota -- by a number of important genetic differences, and this ancient split is strongly supported by phylogenetic analysis. The recently described hyperthermophile Nanoarchaeum equitans, harboring the smallest cellular genome ever sequenced (480 kb), has been suggested as the representative of a new phylum -- the Nanoarchaeota -- that would have diverged before the Crenarchaeota/Euryarchaeota split. Confirming the phylogenetic position of N. equitans is thus crucial for deciphering the history of the archaeal domain.
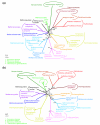
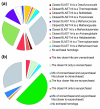
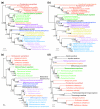
Results: We tested the placement of N. equitans in the archaeal phylogeny using a large dataset of concatenated ribosomal proteins from 25 archaeal genomes. We indicate that the placement of N. equitans in archaeal phylogenies on the basis of ribosomal protein concatenation may be strongly biased by the coupled effect of its above-average evolutionary rate and lateral gene transfers. Indeed, we show that different subsets of ribosomal proteins harbor a conflicting phylogenetic signal for the placement of N. equitans. A BLASTP-based survey of the phylogenetic pattern of all open reading frames (ORFs) in the genome of N. equitans revealed a surprisingly high fraction of close hits with Euryarchaeota, notably Thermococcales. Strikingly, a specific affinity of N. equitans and Thermococcales was strongly supported by phylogenies based on a subset of ribosomal proteins, and on a number of unrelated molecular markers.
Conclusion: We suggest that N. equitans may more probably be the representative of a fast-evolving euryarchaeal lineage (possibly related to Thermococcales) than the representative of a novel and early diverging archaeal phylum.
Figures
Similar articles
-
Gene transfers from nanoarchaeota to an ancestor of diplomonads and parabasalids.Mol Biol Evol. 2005 Jan;22(1):85-90. doi: 10.1093/molbev/msh254. Epub 2004 Sep 8. Mol Biol Evol. 2005. PMID: 15356278
-
The basal phylogenetic position of Nanoarchaeum equitans (Nanoarchaeota).Front Biosci. 2008 May 1;13:6886-92. doi: 10.2741/3196. Front Biosci. 2008. PMID: 18508702
-
Phylogenomic analysis of proteins that are distinctive of Archaea and its main subgroups and the origin of methanogenesis.BMC Genomics. 2007 Mar 29;8:86. doi: 10.1186/1471-2164-8-86. BMC Genomics. 2007. PMID: 17394648 Free PMC article.
-
Mesophilic Crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota.Nat Rev Microbiol. 2008 Mar;6(3):245-52. doi: 10.1038/nrmicro1852. Nat Rev Microbiol. 2008. PMID: 18274537 Review.
-
The origin and evolution of Archaea: a state of the art.Philos Trans R Soc Lond B Biol Sci. 2006 Jun 29;361(1470):1007-22. doi: 10.1098/rstb.2006.1841. Philos Trans R Soc Lond B Biol Sci. 2006. PMID: 16754611 Free PMC article. Review.
Cited by
-
The Archaeal Transcription Termination Factor aCPSF1 is a Robust Phylogenetic Marker for Archaeal Taxonomy.Microbiol Spectr. 2021 Dec 22;9(3):e0153921. doi: 10.1128/spectrum.01539-21. Epub 2021 Dec 8. Microbiol Spectr. 2021. PMID: 34878325 Free PMC article.
-
A Novel Archaeal Lineage in Boiling Hot Springs around Oyasukyo Gorge (Akita, Japan).Microbes Environ. 2021;36(4):ME21048. doi: 10.1264/jsme2.ME21048. Microbes Environ. 2021. PMID: 34819404 Free PMC article.
-
Phylogenetic Signal, Congruence, and Uncertainty across Bacteria and Archaea.Mol Biol Evol. 2021 Dec 9;38(12):5514-5527. doi: 10.1093/molbev/msab254. Mol Biol Evol. 2021. PMID: 34436605 Free PMC article.
-
An Efficient, Nonphylogenetic Method for Detecting Genes Sharing Evolutionary Signals in Phylogenomic Data Sets.Genome Biol Evol. 2021 Sep 1;13(9):evab187. doi: 10.1093/gbe/evab187. Genome Biol Evol. 2021. PMID: 34390574 Free PMC article.
-
Evolution and origin of sliding clamp in bacteria, archaea and eukarya.PLoS One. 2021 Aug 11;16(8):e0241093. doi: 10.1371/journal.pone.0241093. eCollection 2021. PLoS One. 2021. PMID: 34379636 Free PMC article.
References
-
- NCBI Taxonomy Database http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources


