Expression and functional roles of the two distinct NDH-1 complexes and the carbon acquisition complex NdhD3/NdhF3/CupA/Sll1735 in Synechocystis sp PCC 6803
- PMID: 15548742
- PMCID: PMC535876
- DOI: 10.1105/tpc.104.026526
Expression and functional roles of the two distinct NDH-1 complexes and the carbon acquisition complex NdhD3/NdhF3/CupA/Sll1735 in Synechocystis sp PCC 6803
Abstract
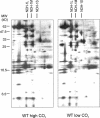
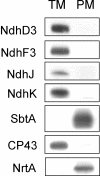
To investigate the (co)expression, interaction, and membrane location of multifunctional NAD(P)H dehydrogenase type 1 (NDH-1) complexes and their involvement in carbon acquisition, cyclic photosystem I, and respiration, we grew the wild type and specific ndh gene knockout mutants of Synechocystis sp PCC 6803 under different CO2 and pH conditions, followed by a proteome analysis of their membrane protein complexes. Typical NDH-1 complexes were represented by NDH-1L (large) and NDH-1M (medium size), located in the thylakoid membrane. The NDH-1L complex, missing from the DeltaNdhD1/D2 mutant, was a prerequisite for photoheterotrophic growth and thus apparently involved in cellular respiration. The amount of NDH-1M and the rate of P700+ rereduction in darkness in the DeltaNdhD1/D2 mutant grown at low CO2 were similar to those in the wild type, whereas in the M55 mutant (DeltaNdhB), lacking both NDH-1L and NDH-1M, the rate of P700+ rereduction was very slow. The NDH-1S (small) complex, localized to the thylakoid membrane and composed of only NdhD3, NdhF3, CupA, and Sll1735, was strongly induced at low CO2 in the wild type as well as in DeltaNdhD1/D2 and M55. In contrast with the wild type and DeltaNdhD1/D2, which show normal CO2 uptake, M55 is unable to take up CO2 even when the NDH-1S complex is present. Conversely, the DeltaNdhD3/D4 mutant, also unable to take up CO2, lacked NDH-1S but exhibited wild-type levels of NDH-1M at low CO2. These results demonstrate that both NDH-1S and NDH-1M are essential for CO2 uptake and that NDH-1M is a functional complex. We also show that the Na+/HCO3- transporter (SbtA complex) is located in the plasma membrane and is strongly induced in the wild type and mutants at low CO2.
Figures

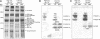





Similar articles
-
Active NDH-1 complexes from the cyanobacterium Synechocystis sp. strain PCC 6803.Plant Cell Physiol. 2006 Oct;47(10):1432-6. doi: 10.1093/pcp/pcl008. Epub 2006 Sep 16. Plant Cell Physiol. 2006. PMID: 16980703
-
Towards functional proteomics of membrane protein complexes in Synechocystis sp. PCC 6803.Plant Physiol. 2004 Jan;134(1):470-81. doi: 10.1104/pp.103.032326. Plant Physiol. 2004. PMID: 14730074 Free PMC article.
-
NdhV subunit regulates the activity of type-1 NAD(P)H dehydrogenase under high light conditions in cyanobacterium Synechocystis sp. PCC 6803.Sci Rep. 2016 Jun 22;6:28361. doi: 10.1038/srep28361. Sci Rep. 2016. PMID: 27329499 Free PMC article.
-
Cyanobacterial NADPH dehydrogenase complexes.Photosynth Res. 2007 Jul-Sep;93(1-3):69-77. doi: 10.1007/s11120-006-9128-y. Epub 2007 Feb 6. Photosynth Res. 2007. PMID: 17279442 Review.
-
Cyanobacterial NDH-1 complexes: multiplicity in function and subunit composition.Physiol Plant. 2007 Sep;131(1):22-32. doi: 10.1111/j.1399-3054.2007.00929.x. Physiol Plant. 2007. PMID: 18251921 Review.
Cited by
-
CupAR negatively controls the key protein CupA in the carbon acquisition complex NDH-1MS in Synechocystis sp. PCC 6803.J Biol Chem. 2024 Sep;300(9):107716. doi: 10.1016/j.jbc.2024.107716. Epub 2024 Aug 22. J Biol Chem. 2024. PMID: 39181331 Free PMC article.
-
Interplay between photosynthetic electron flux and organic carbon sinks in sucrose-excreting Synechocystis sp. PCC 6803 revealed by omics approaches.Microb Cell Fact. 2024 Jul 1;23(1):188. doi: 10.1186/s12934-024-02462-6. Microb Cell Fact. 2024. PMID: 38951789 Free PMC article.
-
CRISPR interference screens reveal growth-robustness tradeoffs in Synechocystis sp. PCC 6803 across growth conditions.Plant Cell. 2023 Oct 30;35(11):3937-3956. doi: 10.1093/plcell/koad208. Plant Cell. 2023. PMID: 37494719 Free PMC article.
-
Deep Proteogenomics of a Photosynthetic Cyanobacterium.J Proteome Res. 2023 Jun 2;22(6):1969-1983. doi: 10.1021/acs.jproteome.3c00065. Epub 2023 May 5. J Proteome Res. 2023. PMID: 37146978 Free PMC article.
-
Honoring two stalwarts of photosynthesis research: Eva-Mari Aro and Govindjee.Photosynth Res. 2023 Jul;157(1):43-51. doi: 10.1007/s11120-022-00988-7. Epub 2023 Feb 27. Photosynth Res. 2023. PMID: 36847891
References
-
- Appel, J., Phunpruch, S., Steinmuller, K., and Schultz, R. (2000). The bi-directional hydrogenase of Synechocystis sp. PCC 6803 works as an electron valve during photosynthesis. Arch. Microbiol. 173, 333–338. - PubMed
-
- Aro, E.-M., Suorsa, M., Rokka, A., Allahverdiyeva, Y., Paakkarinen, V., Saleem, A., Battchikova, N., and Rintamäki, E. (2005). Dynamics of photosystem II: A proteomic approach to thylakoid protein complexes. J. Exp. Bot., in press. - PubMed
-
- Badger, M.R., and Spalding, M.H. (2000). CO2 acquisition, concentration and fixation in cyanobacteria and algae. In Advances in Photosynthesis: Physiology and Metabolism, Vol. 9, R.C. Leegood, T.D. Sharkey, and S. von Caemmerer, eds (Dordrecht: Kluwer Acadademic Publishers), pp. 399–434.
-
- Blum, H., Beier, H., and Gross, J.H. (1987). Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis 8, 93–99.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases


