Abstract
Botanists have long identified bilaterally symmetrical (zygomorphic) flowers with more specialized pollination interactions than radially symmetrical (actinomorphic) flowers. Zygomorphic flowers facilitate more precise contact with pollinators, guide pollinator behaviour and exclude less effective pollinators. However, whether zygomorphic flowers are actually visited by a smaller subset of available pollinator species has not been broadly evaluated. We compiled 53 609 floral visitation records in 159 communities and classified the plants' floral symmetry. Globally and within individual communities, plants with zygomorphic flowers are indeed visited by fewer species. At the same time, zygomorphic flowers share a somewhat larger proportion of their visitor species with other co-occurring plants and have particularly high sharing with co-occurring plants that also have zygomorphic flowers. Visitation sub-networks for zygomorphic species also show differences that may arise from reduced visitor diversity, including greater connectance, greater web asymmetry and lower coextinction robustness of both plants and visitor species—but these changes do not necessarily translate to whole plant-visitor communities. These results provide context for widely documented associations between zygomorphy and diversification and imply that species with zygomorphic flowers may face a greater risk of extinction due to pollinator loss.
Keywords: pollination, floral morphology, ecological specialization, interaction networks
1. Introduction
An axiom of pollination ecology is that flowers with bilateral symmetry are more specialized than flowers with radial symmetry [1–4]. ‘Specialized', however, has multiple meanings in evolution and ecology, which are not mutually exclusive. Specialization may refer to a derived character state in a phylogenetic context [1,3], or the degree to which a flower manipulates pollinator behaviour [5], or it may refer to association with a particular set of pollinators (i.e. pollination syndromes; [6,7])—or, finally, it may refer to association with fewer available pollinator species than seen for comparable plant species [8]. Zygomorphic flowers are derived within the angiosperms [9–11], and extensive research examines how floral structure attracts, guides or excludes pollinators [5,6,12–15]. However, data addressing the fourth sense in which zygomorphic flowers may be specialized—association with fewer pollinators than otherwise expected—are surprisingly sparse.
Floral symmetry has been recognized as an important feature of angiosperm diversity since at least the eighteenth century [4]. Modern treatments identified zygomorphy as derived, and hypothesized that zygomorphic forms facilitate more effective pollination [1,3,16]. Zygomorphy is associated with greater diversification rates [17–19], consistent with the hypothesis that using fewer or more constant pollinators creates more opportunities for reproductive isolation [20]. Greater pollination specialization might also interact with global patterns of diversity, such as latitudinal gradients [21,22]: recent syntheses find evidence that biotic interactions are stronger in the tropics [23,24], though assessments of latitudinal effects on pollination specifically have mixed results [25,26].
Floral symmetry has been considered as an element of pollination syndromes [6,7], but to our knowledge, documentation that zygomorphic flowers associate with fewer pollinator species is restricted to anecdotal observations (e.g. [1,3,5]). Broad confirmation of this understanding would illuminate the research linking pollination to diversification [1,16,18,20,27]. Ecologically, the use of fewer pollinators by zygomorphic flowers may have implications for factors ranging from species' geographic extents to risks of extinction due to pollinator loss.
If floral symmetry creates systematic differences in pollinator associations, these differences should manifest in floral visitation networks. Plant–pollinator associations have been prominent case studies in investigations of ecological networks’ structure and assembly [28–30], geographic variation [25,26] and evolutionary stability [31]. Databases of floral visitation networks have global coverage, recording visitor diversity and sharing among co-occurring angiosperm species across a variety of contexts. Here, we compile a global dataset of floral visitation records to test the hypothesis that zygomorphic flowers have fewer visitor species and examine how this effect may shape plant–pollinator networks.
2. Methods
We compiled floral visitation networks from the Web of Life (www.web-of-life.es) and Interaction Web DataBase (https://iwdb.nceas.ucsb.edu/). Networks varied widely in size, attributable to heterogeneity in the time periods over which they were studied, their observation protocols and the geographic ranges of communities represented (figure 1). We accounted for this in subsequent analyses by simplifying visitation into a binary state (the most common recording mode), and by examining contrasts within individual networks or treating network identity as a random effect (see below).
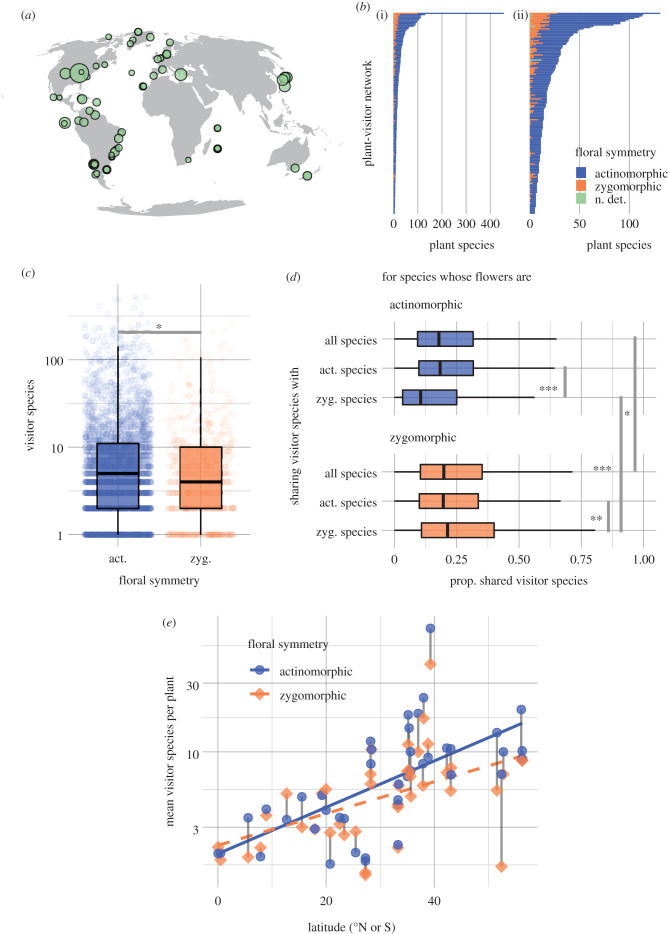
Figure 1.
The global plant-visitation dataset. (a) Global distribution of the plant-visitor networks, with points scaled to indicate numbers of plant taxa. (b) Bar plots giving the number of plant taxa for each network, coloured according to floral symmetry ((i) full dataset, (ii), all networks except for the largest, to provide better visibility). (c) Visitor species per plant, grouped by floral symmetry. (d) Sharing of visitor species with all co-occurring plant species, and co-occurring species with each type of floral symmetry. (e) Mean visitor species per plant in sub-networks for plants with differing symmetries, with grey lines linking points for zygomorphic and actinomorphic taxa from the same network, plotted against latitude of the network location. In (c) and (d), asterisks indicate significant differences in one-tailed Wilcoxon tests: *p < 0.01, **p < 10−4, ***p < 10−5.
We identified unique plant taxa across all networks (hereafter ‘species'; identification of plants and visitors varied in resolution, but 95% of records had plants identified to species), and classified their floral symmetry based on taxonomic knowledge, formal descriptions of species or higher taxa and, when necessary, inspecting images of herbarium sheets or reliably identified fresh flowers. We determined symmetry based on the perianth, but in ambiguous cases also considered symmetry of the androecium or gynoecium; for example, a flower with a very nearly radially symmetric corolla, but stamens arrayed in a bilaterally symmetric manner, would be coded as zygomorphic. In some cases, we classified symmetry not based on individual flowers but on flowering heads (e.g. we considered species in the Asteraceae actinomorphic). We removed species from the working dataset if we were unable to find authoritative descriptions or images, or if they were wind-pollinated (‘n. det.', figure 1b). Data and scripts are posted to Dryad, at https://datadryad.org/10.5061/dryad.gxd2547j3 [32].
We conducted analysis in R v. 4.0 [33]. For each plant species in the dataset, we totalled the visitor species recorded and calculated an index of visitor species sharing, the proportion of visitor species to each plant species that also visit each other co-occurring plant species, averaged across the co-occurring plant species. We calculated sharing with all plants in the same network and sharing with plants in the same network having each type of floral symmetry. We examined the structure of complete networks, and of the sub-networks of visitors to plants with each type of floral symmetry in each community, calculating connectance (the realized proportion of possible plant-visitor links [34]), web asymmetry (the degree to which visitor species outnumber plant species or vice-versa) and coextinction curves (the relationship between species losses in one trophic level and species losses in another [35]) using the networklevel() and second.extinct() functions in the bipartite package [36].
To test for phylogenetic signal in floral symmetry, visitor species count and visitor species sharing, and to control for phylogeny in subsequent analysis, we mapped taxa in our dataset to a recently published time-calibrated supertree of the seed plants (the ‘ALLMB' supertree of [37]), using the congeneric.merge() function from the package pez [38] to add species to the tree if they were not already included. We tested for a phylogenetic signal using the phylosignal package [39], estimating Blomberg's K and K* [40], Abouheif's Cmean [41], Moran's I [42] and Pagel's λ [43] statistics. We performed a principal component analysis of the phylogenetic distance matrix for all plant taxa and used phylogenetic distance principal components as covariates in models fitted to explain variation in visitor count and sharing.
We tested the hypotheses that visitor species count and visitor species sharing differed with respect to floral symmetry by fitting Bayesian multilevel regression models using the brms package [44,45]. Competing models explained visitor count and sharing with a group effect (analogous to a random effect in a ML framework) of source network identity and possible population (i.e. fixed) effects of floral symmetry, latitude and the first two principal components of phylogenetic distance (which jointly explained 61% of variation). We compared model fit in terms of expected log pointwise predictive density (ELPD) using leave-one-out cross-validation, implemented in brms with the LOO() function [46].
3. Results
We compiled 159 networks, recording 53 609 observed visits to 2700 angiosperm species (figure 1a; electronic supplementary material, table S1). We were able to classify floral symmetry for 2685 plant species and were able to place 2582 of these in the time-calibrated supertree [37]. Globally, and in individual networks, zygomorphic flowers were a minority: 498 species (18%) were zygomorphic; only five networks had more zygomorphic than actinomorphic species, while 70 lacked any (figure 1b). Globally, the number of visitor species to zygomorphic flowers was significantly smaller than that for actinomorphic flowers (median four pollinators per zygomorphic species with zygomorphic flowers versus five per actinomorphic species; p = 0.003, one-tailed Wilcoxon test; figure 1c). We found significant phylogenetic signal for floral symmetry (Cmean = 0.85, Moran's I = 0.10, K = 0.20, K* = 0.22, and λ = 0.89; p < 0.001 for all); visitor species count deviated from the null models for Cmean, I, and λ (p < 0.001 for each) but not for K and K*.
At the same time, zygomorphic flowers shared a higher proportion of their visitor species with co-occurring plants (median 0.20 for zygomorphic versus 0.18 for actinomorphic, p = 0.003; figure 1d). Plants also had greater sharing with co-occurring plants of the same floral symmetry (actinomorphic species, median sharing of 0.24 with actinomorphic species versus 0.19 with zygomorphic species, p = 0.003; zygomorphic species, median sharing of 0.30 with zygomorphic species versus 0.27 with actinomorphic species; p < 10−5; figure 1d).
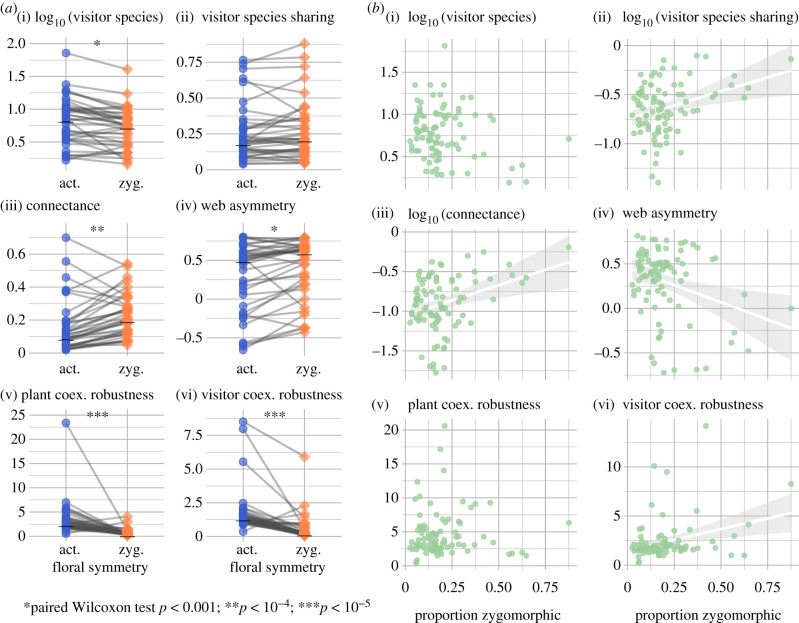
Thirty-nine networks comprising 46 026 visitation records included enough plants of each symmetry type (at least five) to compare sub-networks based on symmetry—that is, to compare networks for plants with different symmetry having access to the same pool of possible visitor species. Zygomorphic sub-networks had significantly fewer mean visitor species per plant (one-tailed paired Wilcoxon test, p < 0.001; figure 2a), but only marginally greater visitor sharing (p = 0.06). Zygomorphic sub-networks had significantly greater connectance (p < 10−4), greater asymmetry (p < 0.001) and lower coextinction robustness, as measured by the exponent of the coextinction curve, for both plants (p < 10−5) and visitors (p < 10−5). Among 89 complete networks that included at least one zygomorphic plant, a higher proportion of zygomorphic plants was associated with greater visitor sharing (figure 2b; product-moment correlation on log-transformed values = 0.22, p = 0.04) and greater connectance (cor = 0.21, p = 0.04), paralleling the sub-network patterns. However, networks with more zygomorphic flowers also had lower asymmetry (cor = −0.26, p = 0.01) and greater visitor coextinction robustness (cor = 0.27, p = 0.01), the opposite of patterns in symmetry-based sub-networks. The proportion of zygomorphic flowers was not correlated with the mean number of visitors per plant or plant coextinction robustness.
Figure 2.
Floral symmetry and plant-visitor network structure. (a) Network descriptive statistics for sub-networks based on floral symmetry. Grey lines link points representing zygomorphic and actinomorphic sub-networks from the same source network, horizontal black bars mark the median value within each floral symmetry type, and asterisks mark significant differences in one-tailed paired Wilcoxon tests. (b) Scatterplots of relationships between the proportion of zygomorphic flowers in a network and the set of network structure metrics in (a); linear regression lines (white with grey 95% CI) are given when the correlation is significant with p ≤ 0.05.
The best-fit model explaining visitor count included floral symmetry, latitude, an interaction between symmetry and latitude, and phylogeny in addition to the group effect of network identity (R2 = 0.28; ΔELPD ≥ 3.8 versus all other models; electronic supplementary material, tables S2 and S3). For visitor sharing, the best-fit model also included all terms; but all simpler models that included an effect of symmetry had ΔELPD ≤ 0.6 (R2 = 0.66 for all such models; electronic supplementary material, tables S4 and S5).
4. Discussion
Zygomorphic flowers have long been considered to be more specialized, which could mean that they are visited by fewer pollinator species. We find that, globally and at the level of individual communities, plants with zygomorphic flowers do indeed have fewer visitor species, and that the visitation networks of plants with zygomorphic symmetry may be structured by this difference (figures 1c,d and 2a). Sub-networks of plants with zygomorphic flowers show greater connectance, greater asymmetry and lower coextinction robustness for both plants and visitors (figure 2a); however, these patterns do not necessarily translate to whole networks (figure 2b). Visitor species count is correlated with latitude north or south (figure 1e), and both floral symmetry and visitor count show significant phylogenetic structure. Bayesian multilevel regressions accounting for these confounding effects nevertheless find significant effects of floral symmetry on visitor species count and sharing.
The visitation records we examine have limitations for assessing pollination specialization. Many of the original studies do not evaluate visitors' pollen transfer and often record visits as a binary, while in reality floral visitors vary considerably in visitation frequency and effectiveness as pollinators. Thus, the effective pollinators of the plants in our dataset are likely a subset of recorded visitors. However, we think it unlikely that data restricted to effective pollinators would reverse the qualitative patterns we see, because a reversal would require systematic bias based on floral symmetry in recording visits.
Our findings that zygomorphic flowers share more visitor species make sense given the other aspects of pollinator specialization associated with zygomorphy. Arithmetically, a plant with fewer visitors can more easily share a high proportion of them even if the absolute number of visitors shared is low, but higher sharing by zygomorphic species probably also reflects network structure. Floral visitation networks are generally nested [30], so the fact that zygomorphic species tend to have fewer visitor species means that their visitors are more likely to interact with many other plant taxa. Zygomorphic flowers, which often manipulate pollinator behaviour, apply pollen to specific parts of pollinators' bodies, or attract pollinators that show greater constancy over a single foraging trip, may be better able to tolerate the elevated risk of receiving heterospecific pollen due to higher visitor sharing [1,20]. The fact that the highest rate of sharing we see in our data is between zygomorphic flowers and other co-occurring zygomorphic species is consistent with this hypothesis.
In our compiled dataset, plants with both types of floral symmetry had more visitor species in communities farther from the equator (figure 1e). This could reflect bias in the resolution of species in more tropical communities, as there are more undescribed species in the tropics and in the Global South [47]. Networks farther from the equator do have more plant and visitor species recorded (cor = 0.54 for plants, cor = 0.65 for visitors, p < 10−5 for both; figure 1a). However, the correlation weakens or disappears if the comparison is with latitude rather than the distance from the equator (i.e. treating distance south of the equator as different from distance north: cor = 0.3, p = 0.69 for plants; cor = 0.16, p = 0.05 for visitors), suggesting this is not simply an issue of better taxonomy in the Global North. Moreover, if plant and visitor species numbers show similar correlations with distance from the equator, we might expect that the number of visitors per plant would be constant across latitudes. An alternative explanation is that floral visitation is more specialized in the tropics, independent of floral symmetry. Testing this hypothesis with greater rigour is beyond the scope of our data, but it would be consistent with recent syntheses finding stronger effects of biological interactions in the tropics [23,24].
An ecological association between floral symmetry and pollinator diversity may explain evolutionary associations, across the angiosperms, between floral zygomorphy and diversification [17,18,19,48]. The direction of the causal relationship, however, remains ambiguous. It may be that association with fewer pollinators creates more opportunities to evolve reproductive isolation [1,20,48] or increases the number of plant species that can be supported by a given community of pollinators [18]; or it may be that more specialized pollination evolves in response to greater diversity as co-occurring species must subdivide available pollinators more finely [27].
Finally, our results have important implications for conservation. We find lower coextinction robustness for sub-networks of plants with zygomorphic flowers, which may be explained by higher connectance and asymmetry of these sub-networks (figure 2a) and higher visitor sharing among zygomorphic flowers (figure 1d). These patterns are not necessarily borne out at the level of whole communities (figure 2b), suggesting that larger communities may be robust to disturbances that endanger their most specialized members. Nevertheless, our analysis does imply that plant taxa with zygomorphic flowers are at greater risk of extinction due to pollinator loss. Despite significant uncertainty in the magnitude of losses, pollinators are widely known to be in rapid decline due to pesticide use, habitat degradation and emerging infections [49,50]. The patterns we find are coarse, but simple rubrics for triage are critical for conserving the more than 300 000 species of angiosperms, most of which will never benefit from individualized conservation assessment. Perhaps more importantly, our results support the idea that ‘compartments' of the global plant–pollinator network must be targets for holistic conservation, focused on preserving interactions and functionality where the network is most fragile [51].
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
J.B.Y. was supported by start-up funds from California State University Northridge, and C.J.C. by a postdoctoral fellowship from the Georgetown Environment Initiative. We thank Shweta Bansal for helpful conversations and Stephen Sondheim for teaching us that a faboid legume can begin an adventure.
Data accessibility
Data including floral symmetry annotations, visitor counts and sharing, and network structure statistics are available with scripts on Dryad Digital Repository: https://datadryad.org/10.5061/dryad.gxd2547j3 [32].
Authors' contributions
J.B.Y. and C.J.C. conceived and designed the study; G.G. annotated floral symmetry with supervision from J.B.Y., and J.B.Y. and G.G. conducted analysis with code provided by C.J.C.; J.B.Y. drafted the paper in consultation with C.J.C. and G.G. All gave final approval for publication and agree to be held accountable for the work performed.
Competing interests
The authors declare no competing interests.
References
- 1.Takhtajan A. 1969. Flowering plants: origin and dispersal. Edinburgh, UK: Oliver & Boyd. [Google Scholar]
- 2.Darwin C. 1877. The various contrivances by which orchids are fertilised by insects, 2nd edn New York, NY: D. Appleton and Company. [Google Scholar]
- 3.Stebbins GL. 1974. Flowering plants: evolution above the species level. Cambridge, MA: Harvard University Press. [Google Scholar]
- 4.Endress PK. 2012. The immense diversity of floral monosymmetry and asymmetry across angiosperms. Bot. Rev. 78, 345–397. ( 10.1007/s12229-012-9106-3) [DOI] [Google Scholar]
- 5.Fenster CB, Armbruster WS, Dudash MR, Wilson P, Fenster CCB, Thomson JD. 2004. Pollination syndromes and floral specialization. Annu. Rev. Ecol. Evol. Syst. 35, 375–403. ( 10.1146/annurev.ecolsys.34.011802.132347) [DOI] [Google Scholar]
- 6.Rosas-Guerrero V, Aguilar R, Martén-Rodríguez S, Ashworth L, Lopezaraiza-Mikel M, Bastida JM, Quesada M. 2014. A quantitative review of pollination syndromes: do floral traits predict effective pollinators? Ecol. Lett. 17, 388–400. ( 10.1111/ele.12224) [DOI] [PubMed] [Google Scholar]
- 7.Ashworth L, Aguilar R, Martén-Rodríguez S, Lopezaraiza-Mikel M, Avila-Sakar G, Rosas-Guerrero V, Quesada M. 2015. Pollination syndromes: a global pattern of convergent evolution driven by the most effective pollinator. In Evolutionary biology: biodiversification from genotype to phenotype, pp. 203–224. Cham, Switzerland: Springer International Publishing. [Google Scholar]
- 8.Poisot T, Bever JD, Nemri A, Thrall PH, Hochberg ME. 2011. A conceptual framework for the evolution of ecological specialisation. Ecol. Lett. 14, 841–851. ( 10.1111/j.1461-0248.2011.01645.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Citerne H, Jabbour F, Nadot S, Damerval C. 2010. The evolution of floral symmetry. Adv. Bot. Res. 54, 85–137. ( 10.1016/S0065-2296(10)54003-5) [DOI] [Google Scholar]
- 10.Sauquet H, et al. 2017. The ancestral flower of angiosperms and its early diversification. Nat. Commun. 8, 16047 ( 10.1038/ncomms16047) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reyes E, Sauquet H, Nadot S. 2016. Perianth symmetry changed at least 199 times in angiosperm evolution. Taxon 65, 945–964. ( 10.12705/655.1) [DOI] [Google Scholar]
- 12.Fenster CB, Armbruster WS, Dudash MR. 2009. Specialization of flowers: is floral orientation an overlooked first step? New Phytol. 183, 502–506. ( 10.1111/j.1469-8137.2009.02852.x) [DOI] [PubMed] [Google Scholar]
- 13.Kampny CM. 1995. Pollination and flower diversity in the Scrophulariaceae. Bot. Rev. 61, 350–366. ( 10.1007/BF02912622) [DOI] [Google Scholar]
- 14.Ushimaru A, Dohzono I, Takami Y, Hyodo F. 2009. Flower orientation enhances pollen transfer in bilaterally symmetrical flowers. Oecologia 160, 667–674. ( 10.1007/s00442-009-1334-9) [DOI] [PubMed] [Google Scholar]
- 15.Macior LW. 2006. Behavioral aspects of coadaptations between flowers and insect pollinators. Ann. Missouri Bot. Gard. 61, 760 ( 10.2307/2395027) [DOI] [Google Scholar]
- 16.Takhtajan A. 1991. Evolutionary trends in flowering plants. New York, NY: Columbia University Press. [Google Scholar]
- 17.Sargent RD. 2004. Floral symmetry affects speciation rates in angiosperms. Proc. R. Soc. B 271, 603–608. ( 10.1098/rspb.2003.2644) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vamosi JC, Vamosi SM. 2010. Key innovations within a geographical context in flowering plants: towards resolving Darwin's abominable mystery. Ecol. Lett. 13, 1270–1279. ( 10.1111/j.1461-0248.2010.01521.x) [DOI] [PubMed] [Google Scholar]
- 19.O'Meara BC, et al. 2016. Non-equilibrium dynamics and floral trait interactions shape extant angiosperm diversity. Proc. R. Soc. B 283, 20152304 ( 10.1098/rspb.2015.2304) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grant V. 1949. Pollination systems as isolating mechanisms in angiosperms. Evolution (NY) 3, 82–97. [DOI] [PubMed] [Google Scholar]
- 21.Mittelbach GG, et al. 2007. Evolution and the latitudinal diversity gradient: speciation, extinction and biogeography. Ecol. Lett. 10, 315–331. ( 10.1111/j.1461-0248.2007.01020.x) [DOI] [PubMed] [Google Scholar]
- 22.Jansson R, Davies TJ. 2008. Global variation in diversification rates of flowering plants: energy vs. climate change. Ecol. Lett. 11, 173–183. ( 10.1111/j.1461-0248.2007.01138.x) [DOI] [PubMed] [Google Scholar]
- 23.Briscoe Runquist R, et al. 2020. Context dependence of local adaptation to abiotic and biotic environments: a quantitative and qualitative synthesis. Am. Nat. 195, 412–431. [DOI] [PubMed] [Google Scholar]
- 24.Hargreaves AL, Germain RM, Bontrager M, Persi J, Amy L. 2020. Local adaptation to biotic interactions: a meta-analysis across latitudes. Am. Nat. 195, 395–411. ( 10.1086/707323) [DOI] [PubMed] [Google Scholar]
- 25.Olesen JM, Jordano P. 2002. Geographic patterns in plant–pollinator mutualistic networks. Ecology 83, 2416–2424. [Google Scholar]
- 26.Ollerton J, Cranmer L. 2002. Latitudinal trends in plant-pollinator interactions: are tropical plants more specialised? Oikos 98, 340–350. ( 10.1034/j.1600-0706.2002.980215.x) [DOI] [Google Scholar]
- 27.Armbruster WS, Muchhala N. 2009. Associations between floral specialization and species diversity: cause, effect, or correlation? Evol. Ecol. 23, 159–179. ( 10.1007/s10682-008-9259-z) [DOI] [Google Scholar]
- 28.Olesen JM, Bascompte J, Dupont YL, Jordano P.. 2007. The modularity of pollination networks. Proc. Natl Acad. Sci. USA 104, 19 891–19 896. ( 10.1073/pnas.0706375104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krishna A, Guimarães PR Jr, Jordano P, Bascompte J. 2008. A neutral-niche theory of nestedness in mutualistic networks. Oikos 117, 1609–1618. [Google Scholar]
- 30.Bascompte J, Jordano P, Olesen JM, Melián CJ, Olesen JM. 2003. The nested assembly of plant–animal mutualistic networks. Proc. Natl Acad. Sci. 100, 9383–9387. ( 10.1073/pnas.1633576100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nuismer SL, Week B, Aizen MA. 2018. Coevolution slows the disassembly of mutualistic networks. Am. Nat. 192, 490–502. ( 10.1086/699218) [DOI] [PubMed] [Google Scholar]
- 32.Yoder JB, Gomez G, Carlson CJ. 2020. Data from: Zygomorphic flowers have fewer potential pollinator species Dryad Digital Repository. ( 10.5061/dryad.gxd2547j3) [DOI] [PMC free article] [PubMed]
- 33.R Core Team. 2020. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing [Google Scholar]
- 34.Dunne JA, Williams RJ, Martinez ND.. 2002. Food-web structure and network theory: the role of connectance and size. Proc. Natl Acad. Sci. USA 99, 12 917–12 922. ( 10.1073/pnas.192407699) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Memmott J, Waser N, Price M. 2004. Tolerance of pollination networks to species extinctions. Proc. R. Soc. B 271, 2605–2611. ( 10.1098/rspb.2004.2909) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dormann CF, Gruber B, Fründ J. 2008. Introducing the bipartite package: analysing ecological networks. R News8, 8–11.
- 37.Smith SA, Brown JW. 2018. Constructing a broadly inclusive seed plant phylogeny. Am. J. Bot. 105, 302–314. ( 10.1002/ajb2.1019) [DOI] [PubMed] [Google Scholar]
- 38.Pearse WD, Cadotte MW, Cavender-Bares J, Ives AR, Tucker CM, Walker SC, Helmus MR. 2015. pez: phylogenetics for the environmental sciences. Bioinformatics 31, 2888–2890. ( 10.1093/bioinformatics/btv277) [DOI] [PubMed] [Google Scholar]
- 39.Keck F, Rimet F, Bouchez A, Franc A. 2016. Phylosignal: an R package to measure, test, and explore the phylogenetic signal. Ecol. Evol. 6, 2774–2780. ( 10.1002/ece3.2051) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blomberg SP, Garland T, Ives AR. 2003. Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution (NY) 57, 717–745. ( 10.1111/j.0014-3820.2003.tb00285.x) [DOI] [PubMed] [Google Scholar]
- 41.Abouheif E. 1999. A method for testing the assumption of phylogenetic independence in comparative data. Evol. Ecol. Res. 1, 895–909. [Google Scholar]
- 42.Gittleman JL, Kot M. 1990. Adaptation: statistics and a null model for estimating phylogenetic effects. Syst. Zool. 39, 227 ( 10.2307/2992183) [DOI] [Google Scholar]
- 43.Pagel M. 1999. Inferring the historical patterns of biological evolution. Nature 401, 877–884. ( 10.1038/44766) [DOI] [PubMed] [Google Scholar]
- 44.Bürkner P-C. 2017. brms: An R package for Bayesian multilevel models using Stan. J. Stat. Softw. 80, 1–28. ( 10.18637/jss.v080.i01) [DOI] [Google Scholar]
- 45.Bürkner P-C. 2018. Advanced Bayesian multilevel modeling with the R package brms. R J. 10, 395 ( 10.32614/RJ-2018-017) [DOI] [Google Scholar]
- 46.Vehtari A, Gelman A, Gabry J. 2017. Practical Bayesian model evaluation using leave-one-out cross-validation and WAIC. Stat. Comput. 27, 1413–1432. ( 10.1007/s11222-016-9696-4) [DOI] [Google Scholar]
- 47.Scheffers BR, Joppa LN, Pimm SL, Laurance WF. 2012. What we know and don't know about Earth's missing biodiversity. Trends Ecol. Evol. 27, 501–510. ( 10.1016/j.tree.2012.05.008) [DOI] [PubMed] [Google Scholar]
- 48.Kay KM, Sargent RD. 2009. The role of animal pollination in plant speciation: integrating ecology, geography, and genetics. Annu. Rev. Ecol. Evol. Syst. 40, 637–656. ( 10.1146/annurev.ecolsys.110308.120310) [DOI] [Google Scholar]
- 49.Lister BC, Garcia A. 2018. Climate-driven declines in arthropod abundance restructure a rainforest food web. Proc. Natl Acad. Sci. USA 115, E10397–E10406. ( 10.1073/pnas.1722477115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Potts SG, Biesmeijer JC, Kremen C, Neumann P, Schweiger O, Kunin WE. 2010. Global pollinator declines: trends, impacts and drivers. Trends Ecol. Evol. 25, 345–353. ( 10.1016/j.tree.2010.01.007) [DOI] [PubMed] [Google Scholar]
- 51.Corbet SA. 2000. Conserving compartments in pollination webs. Conserv. Biol. 14, 1229–1231. ( 10.1046/j.1523-1739.2000.00014.x) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Yoder JB, Gomez G, Carlson CJ. 2020. Data from: Zygomorphic flowers have fewer potential pollinator species Dryad Digital Repository. ( 10.5061/dryad.gxd2547j3) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
Data including floral symmetry annotations, visitor counts and sharing, and network structure statistics are available with scripts on Dryad Digital Repository: https://datadryad.org/10.5061/dryad.gxd2547j3 [32].