Abstract
The phylogenetic relationships of Asian schilbid catfishes of the genera Clupisoma, Ailia, Horabagrus, Laides and Pseudeutropius are poorly understood, especially those of Clupisoma. Herein, we reconstruct the phylogeny of 38 species of catfishes belonging to 28 genera and 14 families using the concatenated mitochondrial genes COI, cytb, and 16S rRNA, as well as the nuclear genes RAG1 and RAG2. The resulting phylogenetic trees consistently place Clupisoma as the sister taxon of Laides, and the five representative Asian schilbid genera form two monophyletic groups with the relationships (Ailia (Laides, Clupisoma)) and (Horabagrus, Pseudeutropius). The so-called “Big Asia” lineage relates distantly to African schilbids. Independent analyses of the mitochondrial and nuclear DNA data yield differing trees for the two Asian schilbid groups. Analyses of the mitochondrial gene data support a sister-group relationship for (Ailia (Laides, Clupisoma)) and the Sisoroidea and a sister-taxon association of (Horabagrus, Pseudeutropius) and the Bagridae. In contrast, analyses of the combined nuclear data indicate (Ailia (Laides, Clupisoma)) to be the sister group to (Horabagrus, Pseudeutropius). Our results indicate that the Horabagridae, recognized by some authors as consisting of Horabagrus, Pseudeutropius and Clupisoma does not include the latter genus. We formally erect a new family, Ailiidae fam. nov. for a monophyletic Asian group comprised of the genera Ailia, Laides and Clupisoma.
Introduction
The family Schilbeidae, one of more than 30 extant families of catfishes, contains five African genera including the type genus Schilbe and five Asian genera, including Clupisoma, Platytropius and Horabagrus [1, 2]. Several morphological phylogenetic studies of the Siluriformes, including those of Mo (1991) [3], De Pinna (1993) [4] and Diogo et al. (2004) [5], evaluated representative genera of Schibidae. Notwithstanding, the phylogenetic relationships of Clupisoma remain unclear because studies other than Mo (1991) [3] did not include both Clupisoma and Platytropius. Mo examined Clupisoma and mentioned Platytropius, but failed to comment on the phylogenetic position of Clupisoma and did not specify which species of Platytropius were examined. Uncertainty exists as to the grouping of genera and the relationship of the Schilbeidae to other catfish families. Molecular phylogenetic analyses by Peng et al. (2005) (based on mitochondrial DNA cytochrome b gene sequences) [6], Hardman (2005) (also using cytochrome b) [7] and Sullivan et al. (2006, 2008) (by using nuclear genes RAG1 and RAG2) [8, 9] all indicated that the Schilbeidae was not monophyletic, and that the analyzed African genera formed a distantly related monophyletic group. The phylogenetic relationships of the five Asian schilbid genera remain uncertain largely due to variation among studies in taxa included, and incomplete sampling of the Asian genera Clupisoma, Pseudeutropius, Ailia, Laides and Horabagrus.
Huang (1981) [10] assigned the species Platytropius sinensis to Platytropius, which originally contained P. siamensis only [11]. Subsequently this species was placed in Clupisoma as C. sinensis by Ng (1999) [12]. Afterwards, Chen et al. (2005) [13] described the new schilbid species Clupisoma nujiangense from China while considering C. sinensis and C. longianalis to be congeners.
Species of Clupisoma are important food catfishes that inhabit the Mekong and upper Salween rivers. In the last two decades, their populations have declined due to over-fishing and anthropogenic habitat changes. Knowledge of the level of genetic diversity of a species can contribute to the understanding of its evolutionary history, and such data are critical for developing effective conservation and management strategies [14]. Genetic diversity may influence the ability of a species to adapt to environmental changes. Thus, such diversity is an important factor in the conservation of endangered species [15].
Herein, we investigate the phylogenetic history of the family Schilbeidae while including representative species of all five Asian genera. Our analyses use the mitochondrial genes COI, cytb, and 16S rRNA, as well as the nuclear genes RAG1 and RAG2. We aim to resolve the groupings of the Asian genera with the inclusion of the Chinese species Clupisoma sinensis.
Materials and Methods
Ethics
All the samples of fishes were bought from local fish dealers in Manzha Market in Menglun Town, Mengla County, Yunnan province, China. (21°56′07.30″N,101°14′56.54″E; elevation: 546m). As food fishes, no permits were required for sampling. All the samples were living in the natural body of water. The housing and husbandry conditions were unclear and all fishes were dead when obtained. Specimens were preserved using 70% ethanol in the Laboratory for Conservation and Utilization of Bio-resources, Yunnan University. All procedures followed corresponding regulations and by-laws and were approved by the Ethics and Experimental Animal Committee of Kunming Institute of Zoology, Chinese Academy of Science, China (KIZ_YP201002).
Sampling and outgroup selection
Seventeen individuals belonging to eight species of six catfish families were sampled (Table 1). Twenty-eight additional sequences from 28 species of 23 genera in 13 catfish families were downloaded from GenBank (Table 1). We used two species each from the Cypriniformes, Clupeiformes and Characiformes as outgroup taxa.
Table 1. The species used in this study and GenBank accession numbers.
| Families name | Genera name | Scientific name | Locality | COI | 16S | Cytb | rag1(1,2) | rag1(3) | rag2 |
|---|---|---|---|---|---|---|---|---|---|
| Sisoridae | Glyptothrax | Glyptothrax lampris 1 | China, Yunnan | JN020065 | JN020051 | JN020080 | JN020106 | JN020091 | JN020122 |
| Glyptothrax lampris 2 | China, Yunnan | JN020066 | JN020052 | JN020081 | JN020107 | JN020092 | JN020123 | ||
| Glyptothrax laosensis | China, Yunnan | JN020067 | JN020053 | JN020082 | JN020108 | JN020093 | JN020124 | ||
| Glyptothrax macromaculatus 1 | China, Yunnan | JN020068 | JN020054 | JN020083 | JN020109 | JN020094 | JN020125 | ||
| Glyptothrax macromaculatus 2 | China, Yunnan | JN020069 | JN020055 | JN020084 | JN020110 | JN020095 | JN020126 | ||
| Glyptothrax macromaculatus 3 | China, Yunnan | JN020070 | JN020054 | JN020085 | JN020111 | JN020095 | JN020127 | ||
| Bagarius yarrelli* | Thailand | EU417766 | AY445910 | DQ119406 | DQ492552 | DQ492446 | DQ492334 | ||
| Pangasiidae | Pangasius | Pangasius beani 1 | China, Yunnan, Menglun | JN020072 | JN020057 | JN020086 | JN020112 | JN020097 | JN020129 |
| Pangasius beani 2 | China, Yunnan, Menglun | JN020073 | JN020058 | JN020087 | JN020113 | JN020098 | JN020130 | ||
| Helicophagus | Helicophagus waandersii* | Thailand | \ | DQ334328 | DQ119468 | DQ492585 | DQ492515 | DQ492402 | |
| Pangasianodon | Pangasianodon hypophthalmus* | Thailand, Nonthabur fish | EF609427 | GU324167 | GQ856796 | DQ492637 | DQ492517 | DQ492404 | |
| Siluridae | Wallago | Wallago attu 1 | Yunnan | JN020076 | JN020061 | AF477828 | JN020115 | JN020100 | JN020133 |
| Wallago attu 2 | Yunnan | JN020076 | JN020061 | AF477828 | JN020116 | JN020101 | JN020134 | ||
| Wallago attu 3 | Yunnan | JN020076 | JN020061 | AF477828 | JN020117 | JN020102 | JN020135 | ||
| Kryptopterus | Kryptopterus minor* | Asia, Aquarium fish trade | \ | AY458879 | AY458895 | DQ492600 | DQ492486 | DQ492373 | |
| Cranoglanidae | Cranoglanis | Cranoglanis bouderius* | China, Guangxi | AY898626 | AY898626 | AY898626 | DQ492572 | DQ492514 | DQ492401 |
| Amblycipitidae | Liobagrus | Liobagrus anguillicauda* | China | EU490878 | AY574353 | AF416888 | EU490965 | EU490983 | EU491002 |
| Liobagrus marginatoides* | China | EU490880 | AY445892 | EU490929 | EU490966 | EU490985 | EU491005 | ||
| Liobagrus marginatus* | China | EU490882 | \ | EU490930 | EU490969 | EU490987 | EU491006 | ||
| Liobagrus sp.* | China | EU490886 | \ | EU490935 | EU490973 | EU490990 | EU491011 | ||
| Schilbidae | Clupisoma ☆ | Clupisoma sinensis 1 | China, Yunnan, Menglun | JN020077 | JN020062 | JN020088 | JN020118 | JN020103 | JN020136 |
| Clupisoma sinensis 2 | China, Yunnan, Menglun | JN020078 | JN020063 | JN020089 | JN020119 | JN020104 | JN020137 | ||
| Clupisoma sinensis 3 | China, Yunnan, Menglun | JN020079 | JN020064 | JN020090 | JN020120 | JN020105 | JN020138 | ||
| Pareutropius | Pareutropius debauwi* | Rep. Congo | NC015837 | NC015837 | NC015837 | DQ492632 | DQ492507 | DQ492394 | |
| Schilbe | Schilbe intermedius* | Rep. Congo | HM882935 | \ | AJ245638 | DQ492615 | DQ492508 | DQ492395 | |
| Ailia+Laides | Ailia | Ailia coila | JN628886 | GQ411080 | EU490901 | DQ492541 | DQ492452 | DQ492340 | |
| Laides | Laides hexanema | EU490866 | \ | EU490915 | DQ492601 | DQ492453 | DQ492341 | ||
| Horabagidae# | Horabagrus | Horabagrus brachysoma* | India | EU490864 | HM579855 | EU490913 | DQ492593 | DQ492554 | DQ492342 |
| Pseudeutropius | Pseudeutropius brachypopterus* | Sumatra, Batang Hari basin | EU490871 | \ | EU490920 | DQ492624 | DQ492455 | DQ492343 | |
| Clariidae | Clarias | Clarias fuscus | China, Yunnan | JN020071 | JN020056 | AF416885 | JN020121 | JN020096 | JN020128 |
| Clarias batrachus* | Thailand, Chao Phraya basin | EF609334 | GQ402540 | DQ119486 | DQ492568 | DQ492521 | DQ492408 | ||
| Clarias gabonensis* | Gabon | HM882915 | \ | \ | DQ492569 | DQ492519 | DQ492406 | ||
| Akysidae | Acrochordonichthys | Acrochordonichthys rugosus* | Thailand | DQ508027 | \ | EU490899 | DQ492539 | DQ492444 | DQ492332 |
| Akysis | Akysis sp.* | Thailand | EU490853 | \ | EU490902 | DQ492542 | DQ492445 | DQ492333 | |
| Akysis parshadi* | China | EU490854 | \ | EU490903 | EU490960 | EU490978 | EU490998 | ||
| Breitensteinia | Breitensteinia cessator* | China | EU490851 | \ | EU490900 | EU490959 | EU490977 | DQ508040 | |
| Bagridae | Mystus | Mystus nemurus 1 | Yunnan | JN020074 | JN020059 | AF499600* | JN020114 | JN020099 | JN020131 |
| Mystus nemurus 2 | Yunnan | JN020075 | JN020060 | AF499600* | JN020114 | JN020099 | JN020132 | ||
| Mystus bocourti* | Thailand | EU490863 | JQ248058 | EU490912 | DQ492589 | DQ492462 | DQ492350 | ||
| Hemibagrus | Hemibagrus wyckioides* | Thailand, Mekong basin | EU490862 | JQ248063 | EU490911 | DQ492587 | DQ492461 | DQ492349 | |
| Leiocassis | Leiocassis poecilopterus* | Sumatra, Batang Hari basin | EU490867 | \ | EU490916 | DQ492603 | DQ492457 | DQ492345 | |
| Anchariidae | Gogo | Gogo arcuatus* | Madagasear, Andriam bombo River | \ | FJ013191 | FJ0131601 | DQ492582 | DQ492528 | DQ492415 |
| Ariidae | Cephalocassis | Cephalocassis borneensis* | Thailand, Chao Phraya basin | \ | FJ626071 | FJ626200 | DQ492563 | DQ192525 | DQ492412 |
| Bagre | Bagre marinus* | USA | GU225559 | DQ990627 | AJ581355 | DQ492553 | DQ492524 | DQ492411 | |
| Ictaluridae | Noturus | Noturus insignis* | USA, NewYork | JN027812 | AY458875 | AY327303 | DQ492639 | DQ492513 | DQ492400 |
| Pylodictis | Pylodictis olivaris* | USA, Pennsylyania | EU525113 | AY458871 | AF484161 | DQ492619 | DQ492512 | DQ492399 | |
| Heteropneustidae | Heteropneustes | Heteropneustes fossilis* | Aquarium fish trade | HQ009491 | FJ432687 | DQ119383 | DQ492591 | DQ492522 | DQ492409 |
| Characiformes | Leporinus | Leporinus fasciatus | \ | HQ17132 | HQ289610 | \ | HQ289223 | HQ289417 | |
| Piabina | Piabina argentea | HM405183 | HQ171283 | GU908175 | \ | HQ289187 | HQ289380 | ||
| Cypriniformes | Cyprinus | Cyprinus carpio* | NC001606 | NC001606 | NC001606 | AY787040 | AY787040 | AY787041 | |
| Danio | Danio rerio* | NC002333 | NC002333 | NC002333 | NM131389 | NM131389 | U71094 | ||
| Clupeifromes | Alosa | Alosa sapidissima* | NC014690 | NC014690 | NC014690 | \ | DQ912116 | DQ912150 | |
| Clupea | Clupea pallasii* | AP009134 | AP009134 | AP009134 | \ | DQ912118 | DQ912152 |
DNA Extraction, PCR and Sequencing
Primers were either designed based on sequences of Pangasiidae retrieved from GenBank by using Primer Premier 5.0 software (Premier Biosoft International), or they were adapted from literature (Table 2). Genomic DNA was isolated from tissue samples by standard phenol/chloroform extraction. PCR were performed in a 30μl reaction mixture containing 20–50 ng templates DNA, 1.2μM dNTP, 0.5μM of the forward and reverse primers, 0.15 units of EX-Taq DNA polymerase enzymes (TaKaRa) and 3 μl of 10× EX-Taq buffer. The amplification reaction was performed using 33 cycles of 30sec at 95°C, annealing at 66 to 55°C for 30sec, and extension of 72°C for 90sec, with an initial step of 4min at 95°C and a final step of 7min at 72°C. PCR products were purified on agarose gels and extracted (Watson BioMedical Inc. Shanghai) and sequenced with a BigDye DNA sequencing kit (ABI) on a 3730XL sequencer (ABI). The sequences were deposited in GenBank (accession numbers listed in Table 1).
Table 2. The primers for PCR amplification and sequencing.
| Gene Fragment | Primer sequences (5'→3') | Source | ||
|---|---|---|---|---|
| COI | F1 | TGT AAA ACG ACG GCC AGT ATT CAA CCA ATC ATA AAG ATA TTG G | amplification | Ivanova(2007) |
| R1 | CAG GAA ACA GCT ATG ACT AAA CTT CTG GAT GTC CAA AAA ATC A | |||
| F1d | TGT AAA ACG ACG GCC AGT TCT CAA CCA ACC ACA ARG AYA TYG G | |||
| R1d | CAG GAA ACA GCT ATG ACT AGA CTT CTG GGT GGC CRA ARA AYC A | |||
| M13F | TGT AAA ACG ACG GCC AGT | Sequencing | Ivanova(2007) | |
| M13R | CAG GAA ACA GCT ATG AC | |||
| 16S | R | CGC CTG TTT AAC AAA AAC AT | amplification and Sequencing | Palumbi (1991) |
| F | CCG GTC TGA ACT CAG ATC ATG T | |||
| Cytb | L14724 | GAC TTG AAA AAC CAC CGT TG | amplification | Xiao (2001) |
| H15915 | CTC CGA TCT CCG GAT TAC AAG AC | |||
| L15138 | ATR ATR ACC GCC TCC GTY GGY TA | Sequencing | Xiao (2001) | |
| L15519 | GGA GAC CCA GAA AAC TTY ACY CC | |||
| H15287 | AGT GGA AGT CGA AGA ATC GTG | |||
| H15560 | GCR TAG GCA AAY AGG AAR TAT C | |||
| Rag1(5') | U69 | TGT TYC TGG CAG CAT TAT GAA | amplification | |
| L1410 | TGY TTC TGM GCC CTT CGT | |||
| U558 | CTT CTA GRT GGC CTG AYG T | Sequencing | ||
| U989 | GAW TTY CCA AAA GAY TTT G | |||
| L594 | TTA AAY ACK TTK AGG ATG ACR T | |||
| L1018 | AAT KGC ACT RAC AAA RTC TTT T | |||
| Rag1(3') | U47 | TTC TTC CKG GST TCC ATC AAT TTG A | amplification | |
| L1423 | TGT TYC CAG ATT CRT TCC CT | |||
| U492 | GTG YCT CAT GTT YGT GGA T | Sequencing | ||
| U903 | TGC CTT GCA CTG TGA CAT TGG CA | |||
| L501 | CAT GAG RCA CAG WGG CCT RC | |||
| L928 | CAT TGC CAA TRT CAC AGT GC | |||
| Rag2 | mhf1 | TGY TAT CTC CCA CCT CTG CGY TAC C | Amplification and Sequencing | Hardman (2004) |
| mhr1 | TCA TCC TCC TCA TCK TCC TCW TTG TA |
The PCR amplification primers and sequencing primers of the nuclear genes were designed based on RAG sequences of Pangasiidae in GenBank.
Sequence analysis
De novo sequences were checked using BLAST [20] against the NCBI database (http://www.ncbi.nlm.nih.gov) to assess sequence similarity. They were aligned using ClustalX 1.83 and manually verified. DAMBE 4.1.19 [16] was used to identify unique haplotypes.
Phylogeny construction
Phylogenies were constructed using maximum likelihood (ML) via RAxML [17], Bayesian inference (BI) executed with MrBaves 3.2 [18], and maximum parsimony (MP) implemented in PAUP* 4.0b10 [19]. We selected the best-fitting models for ML and BI using the Akaike Information Criterion (AIC) [20, 21] as implemented in jModelTest 0.1.1 [22, 23]. BI analysis used four independent MCMC chains run simultaneously for 5 million generations while sampling one tree per 500 replicates, Burnin = 0, and Burninfrac = 0.10, 0.20, 0.30, 0.40, and 0.50. Two runs were conducted independently and the sampled trees were used to construct a 50% majority rule consensus tree after discarding the first 10% as burnin. Bayesian posterior probabilities (BPP), the frequencies of nodal resolution, were mapped on the BI tree. For MP and ML, nodal support was assessed using nonparametric bootstrap sampling [24] of 1000 pseudoreplicates.
Testing tree incongruence
The incongruence among different tree topologies was evaluated using the Approximately Unbiased (AU) test [25], as implemented in the CONSELV0.1i with default scaling and replicate values [26]. Site-wise log-likelihood values were estimated by PAUP*.
Results
MtDNA
The concatenated mtDNA dataset comprised 2300 aligned sites: 626 from the COI fragment, 1137 from cytb, and 537 from the 16S rRNA fragment. The genes consisted of 41 unique haplotypes for 43 sequences of COI, 43 unique haplotypes among 46 sequences of cytb, and 33 unique haplotypes among 36 sequences of 16S rRNA. The combined alignment comprised 2300 positions, of which 941 (40.9%) were potentially parsimony-informative (Table 3).
Table 3. Summary statistics for the genes used in this study.
| COI | 16S | cytb | RAG1(exon1,2) | RAG1(exon3) | RAG2 | |
|---|---|---|---|---|---|---|
| Aligned sites | 626 | 537 | 1137 | 1430 | 1375 | 945 |
| A% (average) | 25.6 | 31.4 | 28.6 | 29.8 | 26.9 | 25.6 |
| G% (average) | 18.7 | 22.6 | 13.9 | 22.3 | 26.3 | 24.7 |
| C% (average) | 27.1 | 24.3 | 29.1 | 22.7 | 22.1 | 25.7 |
| T% (average) | 28.6 | 21.7 | 28.5 | 25.1 | 24.7 | 24.5 |
| Variable sites | 272 (43%) | 206 (39%) | 589 (52%) | 948 (66%) | 675 (49%) | 576 (61%) |
| Parsimony-informative sites | 245 (39%) | 160 (30%) | 536 (47%) | 795 (56%) | 577 (42%) | 472 (50%) |
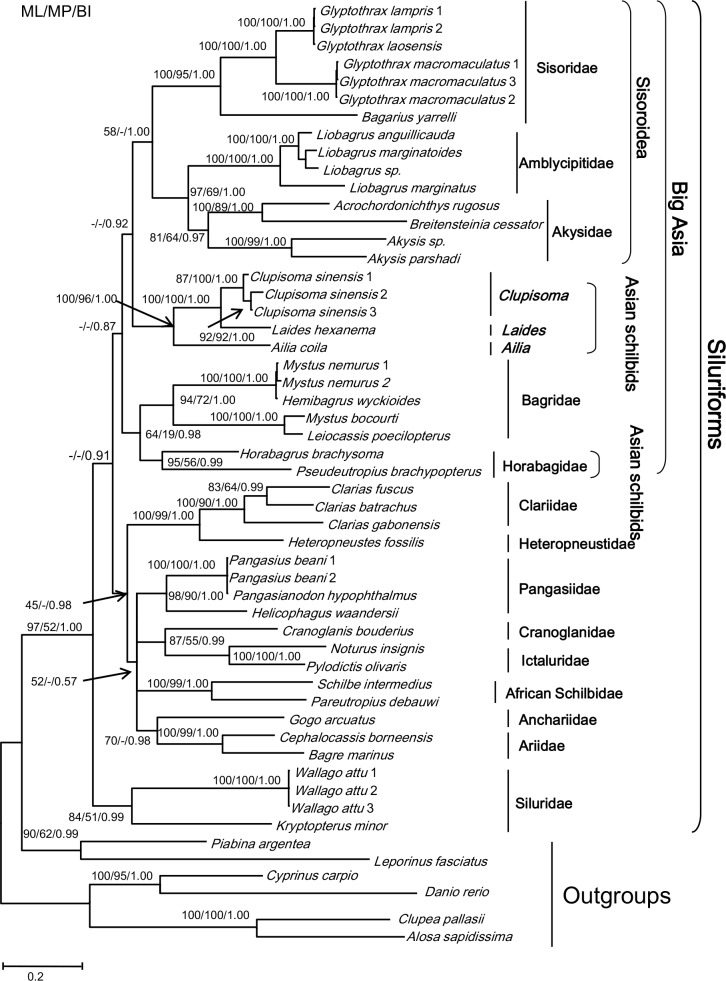
Individual mtDNA gene analyses produced inconsistent topologies with low levels of nodal support, probably due to limited information harbored in a single gene. The trees constructed by analyses of the concatenated data using ML, MP and BI (Fig 1) were consistent for well supported nodes. The five Asian schilbid genera formed two monophyletic groups, one consisting of Clupisoma, Lades and Ailia (BI BPP = 1.0, ML BS = 100% and MP BS = 96%) and the other comprising Horabagrus and Pseudeutropius (BI BPP = 0.99, ML BS = 95% and MP BS = 56%). Clupisoma formed the sister taxon of Laides (BI BPP = 1.0, ML BS = 100% and MP BS = 100%). Excluding the MP tree, the two Asian schilbid groups rooted within the Bagridae. The superfamily Sisoridae, excluding the Aspredinidae, constituted a lineage referred to as “Big Asia” by Sullivan et al. (2006, 2008) [8, 9]. Within “Big Asia”, (Ailia (Laides, Clupisoma)) was the sister-group of the Sisoroidea (BI = 92%), while (Horabagrus, Pseudeutropius) was the sister taxon of the Bagridae (BI = 98%) (Fig 1).
Fig 1. The matrilineal genealogy of the Chinese Clupisoma (as Platytropius) (Schilbeidae) and Pseudeutropius (Pangasiidae) in the Siluriformes derived from the combined mtDNA datasets using ML, MP and BI methods.
Nodal support values are indicated on the branches. The names Sisoroidea and “Big Asia” are after Sullivan et al. (2006) [8].
NuDNA
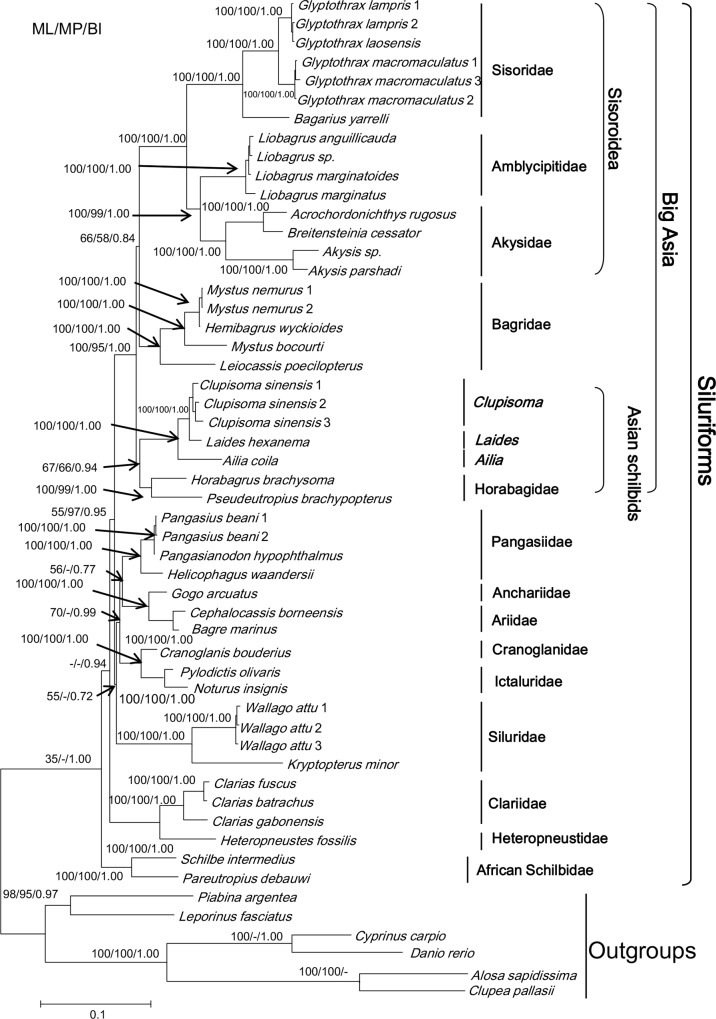
The combined alignment of the nuclear genes RAG1 and RAG2 contained 3750 positions: 1430 from the RAG1 exon 1, 2), 1375 from the RAG1 exon 3, and 945 from RAG2 (Table 3). Among these, 1844 sites (49.3%) were potentially parsimony-informative (Table 3). The tree (Fig 2) displayed likelihood bootstrap proportions, parsimony bootstrap proportions and Bayesian posterior probabilities (BPP).
Fig 2. Phylogenetic relationships of the Siluriformes based on ML, MP and BI analysis of the concatenated datasets of nuclear genes.
Nodal support values are indicated on the branch. The names Sisoroidea and “Big Asia” are after Sullivan et al. (2006) [8].
As with the mt-genes tree, the five Asian schilbid genera also showed the strongly supported monophyletic groups (Ailia (Laides, Clupisoma)) (BI BPP = 1.0, ML BS = 100% and MP BS = 100%) and (Horabagrus, Pseutropius) (BI BPP = 1.0, ML BS = 100% and MP = 99%). However, analyses of the nuDNA data consistently united them as sister taxa (BI = 94%, ML = 67% and MP = 66%) and rooted them in “Big Asia” with strong support (BI BPP = 1.0, ML BS = 100% and MP = 95%). Relationships among this group, the Bagridae, and the superfamily Sisoroidea were not well resolved.
Concatenated MtDNA and NuDNA
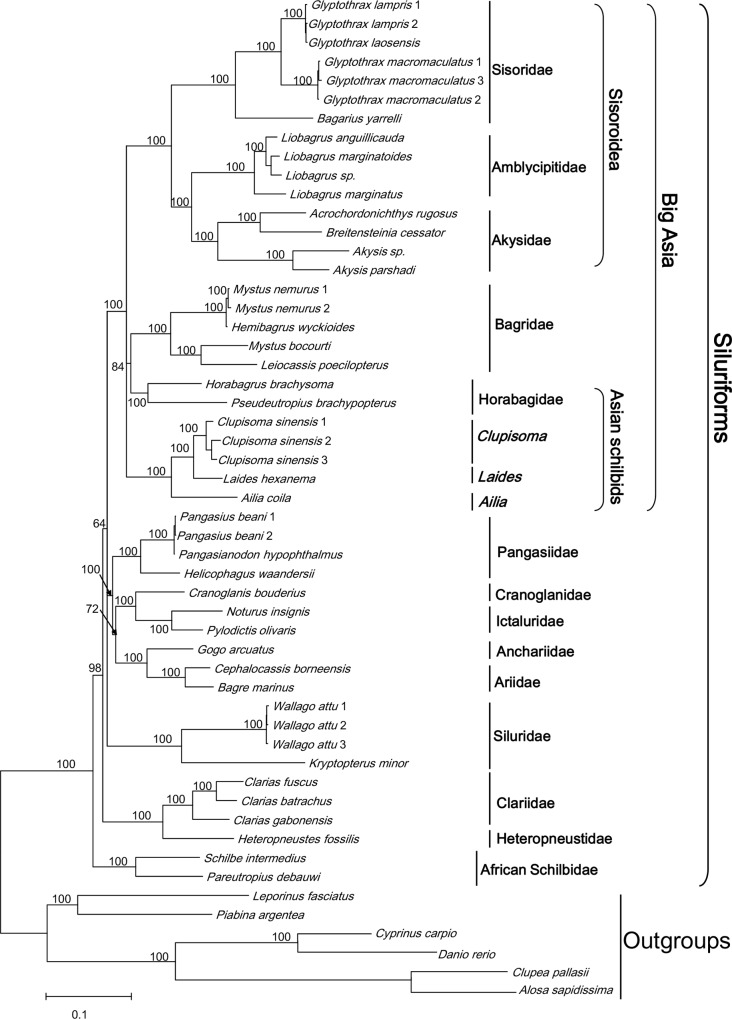
For a total evidence analysis, we have combined three mtDNA genes (COI, 16s and cytb) and two nuclear genes (RAG1 and RAG2). The three mtDNA fragments comprised 2300 aligned sites: 626 from the COI fragment, 537 from the 16S fragment, and 1137 from cytb; and the nuclear dataset consists of 3750 aligned bases: 1430 from the RAG1 (exon 1, 2) fragment, 1375 from the RAG1 (exon 3) fragment and 945 from RAG2 (Table 3). The concatenated datasets were comprised of six fragments including 6050 aligned sites.
The obtained nuDNA trees for the analyzed five Asian schilbid genera (Fig 3; ML and MP trees not shown) were somewhat similar to those of the mt genes trees. Analyses of both genomes resolved two strongly supported monophyletic clades: “Big Asia”, i.e., (Ailia (Laides, Clupisoma)) (BI = 100%, ML = 100% and MP = 100%) and (Horabagrus, Pseudeutropius) (BI = 100%, ML = 100% and MP = 100%). The genomes differed in that the clade (Ailia (Laides, Clupisoma)) did not associate with other taxa in former “Big Asia”. Further, (Horabagrus, Pseudeutropius) had a weakly supported relationship with the family Bagridae.
Fig 3. Phylogenetic relationships of the Siluriformes based on a Bayesian inference analysis of concatenated mtDNA genes and partitioned nuclear genes.
Nodal support values are Bayesian posterior probabilities. The names Sisoroidea and “Big Asia” are after Sullivan et al. (2006) [8].
AU test
The AU test (Table 4) detected significant differences between the mtDNA and nuDNA datasets (P<0.05). Thus, the matrilineal history differed from that of biparental inheritance. We believe this result precluded combining the data sets for phylogenetic analysis inference because each genome had an independent history. However, we retained the result for readers who might be interested in concatenated data results.
Table 4. AU test.
| rank | au | bp | kh | |
|---|---|---|---|---|
| mt BI | 1 | 0.811 | 0.723 | 0.744 |
| nuclear BI | 2 | 0.271 | 0.259 | 0.256 |
| mt+nuclear BI | 3 | 0.023 | 0.018 | 0.02 |
Expanded dataset of Sullivan et al. (2006) [8]
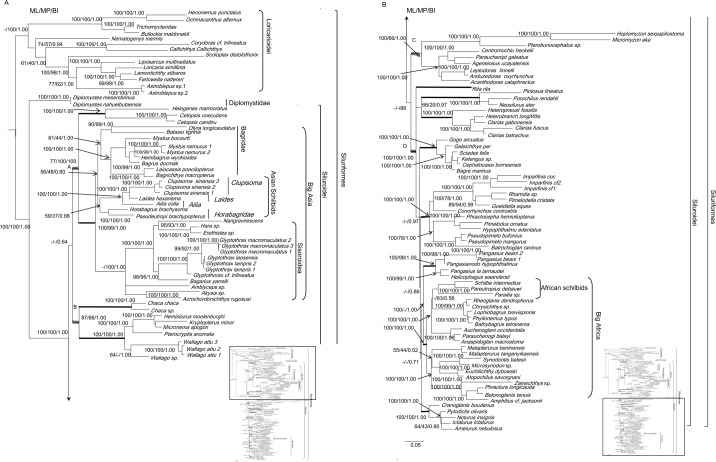
To verify the results from the combined nuDNA dataset, we downloaded the RAG1 and RAG2 sequences of Sullivan et al. (2006) [8] from Siluriformes, to which we added our de novo sequences (Table 1). We reconstructed the ML, MP and BI trees (Fig 4A and 4B). The five Asian schilbid genera remained a monophyletic group with relationship within “Big Asia” shown as ((Aailia (Laides, Clupisoma)), (Horabagrus, Pseudeutropius)). However, this arrangement did not enjoy strong support (BI BPP = 0.88, ML BS = 59%, MP BS = 37%). At higher levels within “Big Asia,” the relationships among the genera in the Bagridae, and the superfamily Sisoroidea were poorly resolved.
Fig 4. Phylogeny of catfishes based on a dataset expanded from Sullivan et al (2006) [8] with nodal support values for BI, ML, and MP, respectively.
The 12 lineages marked by thick branches correspond with those revealed by Sullivan et al. (2006) [8]. (A) Part one of phylogeny of catfishes. The first two clades marked by A, B and ladder-like branch lines are newly resolved herein. (B) Part two of phylogeny of catfishes. Nodal support values are indicated on the branches. The last two clades marked by C, D and ladder-like branches are newly resolved herein.
Analyses of the expanded dataset further resolved relationships within Siluroidei sensu Sullivan et al. (2006) [8]. Their 13 strongly supported monophyletic linages (thick branches in Fig 4A) were recovered along with the further clustering of these groups into major clades(A, B, C and D (Fig 4B). Sullivan et al. (2006) [8] did not obtain interrelationships among their 13 lineages.
Discussion
Phylogeny of Asian schilbid genera
Our analyses consistently support both African and Asian schilbids as monophyletic groups, and show that they are distantly related to one another. Thus, we confirm the non-monophyly of the Schilbeidae as recognized by Peng et al. (2005) [6], Hardman (2005) [7] and Sullivan et al. (2006, 2008) [8, 9].
Recognition of the groups (Ailia (Laides, Clupisoma)) and (Horabagrus, Pseudeutropius) foes not support the monophyly of the so-called “Big Asia” (Figs 1–4) as proposed by Sullivan et al. (2006, 2008) [8, 9]. Analysis of the combined mt gene data and the combined nuclear gene data suggest different suites of relationships among the two groups and other taxa. In the former analysis (Fig 1), the group (Ailia (Laides, Clupisoma)) appears as the sister taxon of the Sisoroidei, and the group (Horabagrus, Pseudeutropius) is the sister taxon of the Bagridae. In contrast, analyses of the combined nuclear data unite the two groups as sister subgroups (Fig 2). Analyses of the expanded dataset of Sullivan et al. (2006) [8] supports this relationship (Fig 4). Because AU testing does not reject either genomic tree, the two results may be equally reliable
Morphological and molecular phylogenetic studies of subsets of the Asian Shilbeidae have been undertaken by Mo (1991) [3], De Pinna (1993) [4], Diogo et al. (2004) [5], Peng et al. (2005) [6], Hardman (2005) [7] and Sullivan et al. (2006, 2008) [8], resulting in differing hypotheses of the relationships among these fishes. This might be in part an artifact of sampling, in particular, the absence of critical taxa. Our study is the first to detail the phylogenetic relationships for all nine recognized genera of Asian schilbids.
In a morphological study, Mo (1991) [3] concluded that the Asian schilbids including Clupisoma comprised two distinct groups: Ailia and the genera Horabagrus, Pseudeutropius and Platytropius. Our results from mtDNA analyses somewhat supports their result by Mo (1991) [3] did not clearly comment on the relationships of Clupisoma or specify which species of Platytropius were examined. He claimed Ailia was associated with the Clariidae and Heteropneustidae while Horabagrus, Pseudeutropius and Platytropius were closer to the Bagridae and Pangasiidae, which differs from our results. We did not have access to De Pinna’s (1993) [4] unpublished dissertation. Thus, we do not know if he examined Clupisoma. Researchers citing his dissertation state that he assigned Horabagrus to its own family because it was distinct from both the Schilbeidae and Bagridae [8]. Further, De Pinna (1993) [4] proposed that all schilbids (including African species) constituted a monophyletic group with the subgroup (Schilbinae (Ailiinae, Laides) being closer to the Pangasiidae than to the Shibeidae (see Fig 2 of Hardman, 2005) [7]. In contrast to our findings, and using a less complete set of Asian schilbids than included in the present study, De Pinna concluded that the Shilbeidae was monophyletic. Diogo et al. (2004) [5] examined Asian Ailia, Laides and Pseudeutropius, and African Schilbe and Siluranodon, and similar to De Pinna obtained results that differed from ours, concluding that the Schilbeidae exclusive of Horabagrus was monophyletic and its sister-group was the Pangasiidae. Unlike Pinna (1993) [4], Diogo et al. (2004) [4] did not propose intergeneric relationships among Ailia, Clupisoma, Horabagrus, Laides and Platytropius.
The molecular phylogenetic studies of Peng et al. (2005) [6] failed to resolve the relationships of Asian schilbids because they sampled Asian Clupisoma only, although they suggested that Chinese schilbids might be closest to either the Bagridae or Siluridae. Hardman (2005) [7] resolved the relationships as (Pseudeutropius (Horabagrus, Clupisoma)) and assigned these genera to the Horabagridae created by De Pinna. However, owing to absence of Ailia and Laides, his study failed to provide an overall phylogenetic scenario of the five genera of Asian schilbids. Further, his resolution of the relationships of Clupisoma differed from ours.
Sullivan et al. (2006, 2008) [8, 9] clustered Ailia with Ladies, and Horabagrus with Pseudeutropius with strong support. Both groups belonged to “Big Asia.” The group (Ailia, Laides) was weakly placed as the sister taxon of the Sisoroidea and the group (Horabagrus, Pseudeutropius) was weakly supported as the sister taxon of Bagridae in their MP and ML trees. Thus, their results are similar to ours based on mtDNA analyses. They could not place Clupisoma owing to its absence in their analyses.
In summary, we propose that 1) the group (Ailia (Laides, Clupisoma)) is monophyletic and 2) its sister-group, based on nuDNA analyses, appears to be (Horabagrus, Pseudeutropius), although this hypothesis conflicts with the matrilineal genealogy based on mtDNA data. Our work specifies the phylogenetic position of Clupisoma, which heretofore was ambiguous, and our hypothesis differs from that of Hardman, which Sullivan et al. (2006, 2008) [8, 9] assumed to be true.
Tree sensitivity
Many factors affect the topologies of phylogenetic trees, including choice of outgroup, ingroup representation, the evolution of genes, long-branch attraction (LBA), and method of tree construction [27]. Two of these factors considerably affect the topologies of the trees for catfishes: choice of genome and taxonomic representation. Phylogenetic relationships based on the mtDNA and nuDNA differ significantly, a discovery termed cytonuclear discordance [28]. The resulting trees differ not only among the members of “Big Asia” but also among other catfishes (Figs 1 and 2). The conflict is not unusual [29, 30]. Our results reinforce the hypothesis that nuclear and mt genes may have different evolutionary trajectories.
The density of ingroup sampling also affects trees. The addition of 17 ingroup sequences (Table 1) to the dataset of Sullivan et al. (2006) [8] changes the topology of the tree greatly. It further resolves the relationships among the 13 lineages comprising the suborder Sisoroidei (Fig 4). Saitoh et al. (2006) [31], Wang et al. (2007) [32], Li et al. (2008) [33], Yang et al. (2010) [34], Telford and Copley (2011) [27] and Wang et al. [35] emphasized the importance of increasing the density of ingroup sampling. The present study provides support for this approach.
Taxonomic implications
Taxonomy should reflect historical relationships [36]. Based on his own analyses and those of Mo (1991) [3], Hardman (2005) [7] recognized the Horabagridae of De Pinna (1993) [4] as containing the genera Horabagrus, Pseudeutropius and Clupisoma. Sullivan et al. (2006) [8] followed this assignment. Our results support the recognition of the Horabagridae vis-à-vis Asian taxa, but with the exclusion of Clupisoma. The Horabagridae De Pinna (1993) [4] contains Horabagrus and Pseudeutropius only. We note that sometimes Horabagrus has been assigned to the Bagridae [3].
Recognition of the Horabagridae renders the Schilbeidae a polyphyletic family. The type genus of Schilbeidae, Schilbe, is native to Africa. Because African schilbids are not the sister group of Asian genera [3], and to obtain a taxonomy that reflects the phyletic history of these Asian catfishes, we formally erect a new family Ailiidae fam. nov. (type genus Ailia) for monophyletic Asian group comprised of the genera Ailia, Laides and Clupisoma. This results in recognition of the following taxonomy for these catfishes:
Class Actinopterygii
Order Siluriformes
Suborder Sisoroidei
Family Horabagridae: Horabagrus (Asia), Pseudeutropius (Asia)
Family Ailiidae fam. nov.: Ailia (Asia), Laides (Asia), Clupisoma (Asia)
Family Schilbeidae: Schilbe (Africa), Irvineia (Africa), Pareutropuis (Africa), Parailia (Africa), Siluranodon (Africa), Platytropius (Asia), Eutropiichthys (Asia),Neotropius (Asia), Proeutropiichthys (Asia), Silonia (Asia) [3]
We do not have any specimens of Horabagridae or Ailiidae, we obtained the morphological information of seven species within these two lineages from FishBase (http://www.fishbase.org/search.php?lang=English). Unfortunately, only one morphological trait was available for all seven species. The total numbers of soft rays of anal fin in Horabagridae ranged from 31 to 33, while the ones in Ailiidae ranged from 39 to 55 (S1 Table). These data are congruent with our hypothesis of a new family of Ailiidae. In addition, images displayed on the website show differences in body shape: the abdominal line of Horabagridae tends to be flat, while those of the Ailiidae curve. These data also show divergence between these two lineages (S1 Fig). The morphological differences correspond with the molecular evidence for a new family.
Undoubtedly, morphological evidence is crucial to propose a new family from within an established family. We encourage the acquisition of deeper morphology evidence or other disciplines to further test our hypothesis of the Ailiidae.
Accession Numbers
All the sequences by this study have been submitted to GenBank. The accession numbers together with the downloaded data were listed in Table 1.
Supporting Information
(PDF)
(DOCX)
Acknowledgments
This work was supported by one grant from National Basic Research Program of China. (973 Program) (No.2007CB411600), URL: http://www.most.gov.cn/cxfw/200806/t20080617_62500.htm, Yaping Zhang contributed to the conceiving and designing of the experiments in our manuscript. This work was also supported by three grants from National Natural Science Foundation of China. The first grant: No.30870291, URL: http://npd.nsfc.gov.cn/projectDetail.action?pid=30870291, Jing Luo contributed to reparation and the writing of the manuscript; the second grant: No.30930071, UTL: http://npd.nsfc.gov.cn/projectDetail.action?pid=30930071, Shaojun Liu contributed to the conceiving and designing of the experiments in our manuscript; the last one grant: No.31272335, UTL: http://npd.nsfc.gov.cn/fundingProjectSearchAction!search.action, Ziming Chen contributed to reparation and the writing of the manuscript.
Data Availability
All the sequences for this study have been submitted to GenBank. The accession numbers together with the downloaded data were listed in Table 1.
Funding Statement
2007CB411600, http://www.most.gov.cn/cxfw/200806/t20080617_62500.htm, 973 program of China, Y.P.Z., contributed to the conceiving and designing of the experiments. 30870291, http://npd.nsfc.gov.cn/projectDetail.action?pid=30870291, National Natural Science Foundation of China, J.L., contributed to reparation and writing of the manuscript. 30930071, http://npd.nsfc.gov.cn/projectDetail.action?pid=30930071, National Natural Science Foundation of China, S.J.L., contributed to conceiving and designing the experiments. 31272335, http://npd.nsfc.gov.cn/fundingProjectSearchAction!search.action, National Natural Science Foundation of China, Z.M.C., contributed to the conceiving and designing of the experiments.
References
- 1.Ferraris J. Checklist of catfishes, recent and fossil (Osteichthyes: Siluriformes), and catalogue of siluriform primary types. Zootaxa. 2007; 1418: (1–628). [Google Scholar]
- 2.Nelson JS. Fishes of the world 4th ed. New York: John Willey and Sons Press; 2006. [Google Scholar]
- 3.Mo TP. Anatomy, relationships and systematics of the Bagridae (Teleostei: Siluroidei) with a hypothesis of siluroid phylogeny. Theses Zool. 1991; 17: 1–216. [Google Scholar]
- 4.Pinna, MCC. Higher-level phylogeny of Siluriformes, with a New Classification of the Order (Teleostei, Ostariophysi). Unpublished Ph.D. Dissertation. New York: The City University of New York; 1993.
- 5.Diogo R, Chardon M, Vandewalle P. Osteology and myology of the cephalic region and pectoral girdle of Schilbe mystus and comparison with other Schilbids, with comments on the monophyly and phylogenetic relationships of the Schilbeidae (Teleostei: Siluriformes). Anim Biol. 2004; 54: 91–110. [Google Scholar]
- 6.Peng ZG, Zhang YG, He SP, Chen YY. Phylogeny of Chinese catfishes inferred from mitochondrial cytochrome b sequences. Acta Genetica Sinica. 2005; 32: 145–154. [PubMed] [Google Scholar]
- 7.Hardman M. The phylogenetic relationships among non-diplomystid catfishes as inferred from mitochondrial cytochrome b sequences; the search for the ictalurid sister taxon (Otophysi: Siluriformes). Mol Phylogenet Evol. 2005; 37: 700–720. [DOI] [PubMed] [Google Scholar]
- 8.Sullivan JP, Lundberg JG, Hardman M. A phylogenetic analysis of the major groups of catfishes (Teleostei: Siluriformes) using RAG1 and RAG2 nuclear gene sequences. Mol Phylogenet Evol. 2006; 41: 636–662. [DOI] [PubMed] [Google Scholar]
- 9.Sullivan JP, Peng ZG, Lundberg JG, Peng JL, He SP. Molecular evidence for diphyly of the Asian catfish family Amblycipitidae (Teleostei: Siluriformes) and exclusion of the South American Aspredinidae from Sisoroidea. Proc Natl Acad Sci Philadelphia. 2008; 157: 51–65. [Google Scholar]
- 10.Huang S. On two new species of the catfish genus Platytropius Hora from Yunnan, China. Acta Zootaxonomica Sinica. 1981; 6: 437–440. [Google Scholar]
- 11.Chu XL, Zhang BS, Dai DY. Fauna Sinica, Osteichthyes, Siluriformes. Beijing: China Science Press; 1999. [Google Scholar]
- 12.Ng HH. Laides longibarbis, a valid species of schilbeid catfish from Indochina (Teleostei: Siluriformes). Ichthyol Explor Freshw. 1999; 10: 381–385. [Google Scholar]
- 13.Chen XY, Ferraris CJ, Yang JX. A new species of catfish of the genus Clupisoma (Siluriformes: Schilbeidae) from the Salween River, Yunnan, China. Copeia. 2005; 566–570. [Google Scholar]
- 14.Vrijenhoek RC. Genetic diversity and fitness in small populations In: Loeschcke V, Tomiuk J, Jian SK, editors. Basel: Birkhauser; In Conservation genetics. 1994. pp. 37–53. [Google Scholar]
- 15.Na-Nakorn U, Sukmanomon S, Nakajima M, Taniguchi N, Kamonrat W, Poompuang S, et al. MtDNA diversity of the critically endangered Mekong giant catfish (Pangasianodon gigas Chevey, 1913) and closely related species: implications for conservation. Anim Conserv. 2006; 9: 483–494. [Google Scholar]
- 16.Xia X, Xie Z. DAMBE: software package for data analysis in molecular biology and evolution. J Hered. 2001; 92: 371–373. [DOI] [PubMed] [Google Scholar]
- 17.Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol. 2008; 57: 758–771. 10.1080/10635150802429642 [DOI] [PubMed] [Google Scholar]
- 18.Huelsenbeck JP, Ronquist F. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics. 2001; 17: 754–755. [DOI] [PubMed] [Google Scholar]
- 19.Swofford DL. PAUP*. Phylogenetic Analysis Using Parsimony (*and other methods). Massachusetts: Sunderland Sinauer Associates Press; 2002. [Google Scholar]
- 20.Goldman N. Simple diagnostic statistical tests of models of DNA substitution. J Mol Evol. 1993; 37: 650–661. [DOI] [PubMed] [Google Scholar]
- 21.Huelsenbeck JP, Crandall KA. Phylogeny estimation and hypothesis testing using maximum likelihood. Ann Rev Ecol Syst. 1997; 28: 437–466. [Google Scholar]
- 22.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003; 52: 696–704. [DOI] [PubMed] [Google Scholar]
- 23.Posada D. jMODELTEST: Phylogenetic model averaging. Mol Phylogent Evol. 2008; 25: 1253–1256. [DOI] [PubMed] [Google Scholar]
- 24.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985; 39: 783–791. [DOI] [PubMed] [Google Scholar]
- 25.Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Syst Biol. 2002; 51: 492–508. [DOI] [PubMed] [Google Scholar]
- 26.Shimodaira H, Hasegawa M. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics. 2001; 17: 1246–1247. [DOI] [PubMed] [Google Scholar]
- 27.Telford MJ, Copley RR. Improving animal phylogenies with genomic data. Trends Genet. 2011; 5: 186–197. [DOI] [PubMed] [Google Scholar]
- 28.Toews DPL and Bresford A. The biogeography of mitochondrial and nuclear discordance in animals. Mol Ecol. 2012; 21: 3907–3930. 10.1111/j.1365-294X.2012.05664.x [DOI] [PubMed] [Google Scholar]
- 29.Yu L, Peng D, Liu J, Luan PT, Liang L, Lee H, et al. On the phylogeny of Mustelidae subfamilies: analysis of seventeen nuclear non-coding loci and mitochondrial complete genomes. BMC Evol Biol. 2011; 11: 1471–2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang XP, Yu L, Roos C, Ting N, Chen CP, Wang J, et al. Phylogenetic relationships among the colobine monkeys revisited: new insights from analyses of complete mt genomes and 44 nuclear non-coding markers. PLoS ONE. 2012; 7: e36274 10.1371/journal.pone.0036274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saitoh K, Sado T, Mayden RL, Hanzawa N, Nakamura K, Nishida M, et al. Mitogenomic evolution and interrelationships of the Cypriniformes (Actinopterygii: Ostariophysi): The first evidence toward resolution of higher-level relationships of the world’s largest freshwater fish clade based on 59 whole mitogenome sequences. J Mol Evol. 2006; 63: 826–841. [DOI] [PubMed] [Google Scholar]
- 32.Wang X, Li J, He S. Molecular evidence for the monophyly of East Asian of Cyprinidae (Teleostei: Cypriniformes) derived from the nuclear recombination activating gene 2 sequences. Mol Phylogenet Evol. 2007; 42: 157–170. [DOI] [PubMed] [Google Scholar]
- 33.Li J, Wang X, Kong X, Zhao K, He S, Mayden RL. Variation pattern of the mitochondrial 16s rRNA gene with secondary structure constrains and their application to phylogeny of Cyprinine fishes (Teleostei: Cypriniformes). Mol Phylogenet Evol. 2008; 47: 472–487. 10.1016/j.ympev.2007.09.012 [DOI] [PubMed] [Google Scholar]
- 34.Yang L, Li H. Yunnan Wetlands Biology, Wetlands vertebrata In: Yunnan Wetlands Beijing, China: China Forestry Publishing House;. 2010. pp. 471–488. [Google Scholar]
- 35.Wang J, Wu XY, Chen ZM, Yue ZhP, Ma W, Chen S.Y, et al. Molecular phylogeny of European and African Barbus and their West Asian relatives in the Cyprininae (Teleostei: Cypriniformes) and orogenesis of the Qinghai-Tibetan Plateau. Chin Sci Bull. 2013; 58: 3738–3746. [Google Scholar]
- 36.Hennig W. Phylogenetic Systematics. Urbana: University of Illinois Press; 1966. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(DOCX)
Data Availability Statement
All the sequences for this study have been submitted to GenBank. The accession numbers together with the downloaded data were listed in Table 1.