Abstract
Rationale
Thoracic aortic aneurysms leading to acute aortic dissections (TAAD) can be inherited in families in an autosomal dominant manner. As part of the spectrum of clinical heterogeneity of familial TAAD, we recently described families with multiple members that had TAAD and intracranial aneurysms or TAAD and intracranial and abdominal aortic aneurysms inherited in an autosomal dominant manner.
Objective
To identify the causative mutation in a large family with autosomal dominant inheritance of TAAD with intracranial and abdominal aortic aneurysms by performing exome sequencing of two distantly related individuals with TAAD and identifying shared rare variants.
Methods and Results
A novel frame shift mutation, p. N218fs (c.652delA), was identified in the SMAD3 gene and segregated with the vascular diseases in this family with a LOD score of 2.52. Sequencing of 181 probands with familial TAAD identified three additional SMAD3 mutations in 4 families, p.R279K (c.836G>A), p.E239K (c.715G>A), and p.A112V (c.235C>T) resulting in a combined LOD score of 5.21. These four mutations were notably absent in 2300 control exomes. SMAD3 mutations were recently described in patients with Aneurysms Osteoarthritis Syndrome and some of the features of this syndrome were identified in individuals in our cohort, but these features were notably absent in many SMAD3 mutation carriers.
Conclusions
SMAD3 mutations are responsible for 2% of familial TAAD. Mutations are found in families with TAAD alone, along with families with TAAD, intracranial aneurysms, aortic and bilateral iliac aneurysms segregating in an autosomal dominant manner.
Keywords: thoracic aortic aneurysm and dissection, intracranial aneurysm, arterial aneurysms, SMAD3
Aneurysms involving the ascending thoracic aorta leading to acute aortic dissections (TAAD) can be inherited in families in an autosomal dominant manner with decreased penetrance and variable expression, termed familial thoracic aortic aneurysms and dissections (FTAAD).1 The phenotypic variability is evident within families and between unrelated families, not only in the age of onset and aortic disease presentation, but also in the presence of other clinical features that segregate with TAAD, which can include congenital defects (e.g., bicuspid aortic valve or patent ductus arteriosus) or vascular diseases elsewhere (e.g., intracranial aneurysms (ICAs) and occlusive disease).2–4 As part of the spectrum of clinical heterogeneity in familial TAAD, we recently described families with multiple members that had TAAD and ICAs or TAAD, ICAs, and abdominal aortic aneurysms (AAAs) inherited in an autosomal dominant manner.5 A majority of these families did not have a mutation in a known gene for familial TAAD, such as the TGF-β receptors type I and II (TGFBR1 and TGFBR2), and smooth muscle aortic α-actin (ACTA2).
TAADs are also a complication of genetic syndromes, such as Marfan syndrome (MFS) or Loeys-Dietz syndrome (LDS), which are caused by mutations in fibrillin-1 (FBN1) and the TGFBR1 and TGFBR2, respectively.6, 7 Although mutations in these genes are associated with syndromic features in addition to TAAD, FTAAD can result from mutations in these genes in the absence of these additional features.8–11 Typically, these families represent the mild end of the disease spectrum when compared with MFS and LDS with a later age of onset of TAAD. Recently, a novel form of TAAD with tortuosity throughout the arterial tree, mild craniofacial features, skeletal and cutaneous anomalies, and early-onset osteoarthritis was described.12 The disorder, termed Aneurysms Osteoarthritis Syndrome (AOS), is due to heterozygous mutations in SMAD3, which encodes a protein involved in downstream cellular signaling initiated by TGF-β binding to its receptors (TGFBR1 and TGFBR2). SMAD3 mutations described in AOS patients include missense and frame shift mutations in exon 6 of the gene.
We sought to identify the causative mutation in a large family, TAA549, with dominant inheritance of TAAD and ICAs, by performing exome sequencing of two distant relatives with TAAD and identifying shared rare variants between these individuals. Using this approach, a novel frame shift mutation was identified in SMAD3 as causing arterial aneurysms in this family. Sequencing of 181 FTAAD probands identified three additional SMAD3 mutations in 4 families, indicating that SMAD3 mutations are responsible for approximately 2% of FTAAD and lead to an inherited predisposition for TAAD, ICAs, and AAAs.
Methods
Family Recruitment and Characterization
The study protocol was approved by the Institutional Review Board of the University of Texas Health Science Center at Houston and the study participants gave informed consent. Families with two or more members affected with TAAD were enrolled into the study. Phenotypic characterization of vascular diseases, including TAAD, ICAs and AAAs, was previously described.5 Blood or saliva samples were obtained from affected individuals and family members. Medical records, including imaging studies of the aorta and cerebral vessels, surgical reports, hospital records, and physicians’ notes, were reviewed. Phenotypic features beyond the vascular system were assessed in eight SMAD3 mutation carriers examined by clinical geneticists. SMAD3 mutation carriers were interviewed concerning joint pain and complaints, and the medical records were reviewed for diagnosis of osteoarthritis. The ethnicity of the 181 FTAAD probands was 86% European American, 5% African American, 1% Asian, 4% Hispanic and 3% other ethnicity. SMAD3 variants were only identified in European Americans.
Targeted capture and massive parallel sequencing
Genomic DNA was extracted from peripheral blood lymphocytes using standard protocols. Five micrograms of DNA from two affected individuals in family TAA549 (1/16 coefficient of relatedness) were used for construction of the shotgun sequencing library as described previously using adaptors for paired-end sequencing.13 Exome sequences were captured by SeqCap EZ. Exome probes version 1.0 (Roche) and recovered according to manufacturer’s directions. Enriched libraries were then sequenced on a Illumina GAIIx using manufacturer protocols.
Read mapping and variant analysis
Reads were mapped to the reference human genome (UCSC hg19) with BWA(Burrows-Wheeler Aligner)14, and variants called with SAMtools.15 Insertion-deletion (indel) variants affecting the coding sequence were identified after a Smith-Waterman realignment of the BWA calls. Single nucleotide variants (SNVs) and indels were filtered to >8× and quality >30. Annotation of variants was performed using the SeattleSeq server (http://gvs.gs.washington.edu/SeattleSeqAnnotation/). The identified variants were then filtered against exome data from 21 non-affected control individuals for indel and SNV calls to identify novel non-synonymous and splice acceptor and donor site variant that was present as heterozygous genotype in both individuals. These variants were considered as a candidate mutation.
Confirmation Sequencing and Linkage Analysis
Bidirectional DNA sequencing of candidate variants were done using primers designed 60–120 base pairs from the variant. Polymerase chain reaction (PCR) amplifications were carried out using HotStar Taq™ DNA polymerase (Qiagen Inc.Valencia, CA). PCR products were treated with EXOSAP-IT (Affymetrix, Inc. OH) to digest the primers and followed with sequencing PCR using the BigDye™ sequencing reaction mix (Applied Biosystems, CA). The sequencing PCR products were purified using the BigDye XTerminator kit (Applied Biosystems, CA) and then loaded on an ABI3730xl sequencing instrument using the Rapid36 run module. DNA sequencing results were analyzed using the Mutation Surveyor software (SoftGenetics, PA). SMAD3 sequencing of all exons and flanking introns were carried out using DNA from 181 probands with FTAAD and SMAD3 mutations were reported based on the RefSeq codes NM_005902.3 (SMAD3 mRNA) and NP_005893.1 (SMAD3 protein). The SMAD3 rare variants identified in family TAA549 and 4 FTAAD probands were not present in 2300 exomes from the Exome Sequencing Project (approximately two-thirds European descent and one-third African descent)16. The mutational status of family members, who carry the mutation but are unaffected and not essential in demonstrating segregation of the mutation with the disease phenotype, are not reported in the pedigrees.
Two-point linkage analysis with candidate variant status was performed in the families with SMAD3 mutations. An affected-only analysis was done with unknown and unaffected individuals both designated as unknown as far as vascular disease status in the analysis. The disease-allele frequency were defined as previously described and 0.001 was the minor allele frequency of the candidate variants.17 Log of odds (LOD) scores were calculated with MLINK program of the computer software FASTLINK, version 3.P.18
Results
The vascular disease in family TAA549 demonstrates autosomal dominant inheritance of a phenotype characterized by presentation of aneurysms involving various arteries, including the thoracic and abdominal aorta, iliac, and intracranial arteries (full phenotypic data of this family was previously reported5; Figure 1A). DNA from two distantly related members (circled in Figure 1A) affected with thoracic aortic disease underwent exome sequencing and approximately 17,000 variants were identified in each individual. Genetic variants were not pursued further if they were present in the NCBI dbSNP database (GRCh37/hg19), 1000 Genomes Project, and in-house controls (21 control exomes). Since the vascular disease in this family showed autosomal dominant inheritance, the analyses focused on novel, heterozygous non-synonymous and insertion/deletion variants present in both affected family members. There were 219 and 271 non-synonymous rare variants, and 4 and 6 frame shift variants in these two individuals but only 11 rare non-synonymous variants and one frame shift mutation were shared (Table 1). This list included a nucleotide deletion in exon 5 (c.652delA) in SMAD3, resulting in a frame shift mutation (p.N218fs) and introducing a premature stop codon. This frame shift mutation was present in all individuals with vascular disease in the family, including TAAD, ICA, AAA, and bilateral iliac aneurysms (Figure 1A). The SMAD3 mutation segregated with vascular diseases in the family with a LOD score of 2.52 and was absent from approximately 2300 control exomes. Therefore, SMAD3 was pursued further as a putative causative gene for FTAAD.
Figure 1. Pedigrees of FTAAD families with SMAD3 mutations.
(A) Pedigree of TAA549 that was used for exome sequencing assay. (B) Additional FTAAD pedigrees with SMAD3 mutations. Filled symbols indicate individuals with arterial aneurysms (black- thoracic aortic aneurysm and/or dissection, red- intracranial aneurysm or subarachnoid hemorrhage, green- abdominal aortic aneurysm, blue- iliac artery aneurysm, gray- sudden death without a known cause). Members of TAA549 who underwent exome sequencing are circled. A # symbol indicates individuals with osteoarthritis or other joint degenerative disease, a * symbol indicates the individual who died of unrelated causes, and a horizontal line above the symbol indicates the individual was examined by a clinical geneticist.
Table 1.
Variants identified by exome sequencing in two affected individuals of family TAA549.
| Gene | Physical location | AA alteration | Alteration/Total AA | PolyPhen-2 analysis |
|---|---|---|---|---|
| SMAD3 | chr15:67462935 | 1 bp deletion | 218/426 | |
| AFAP1L1 | chr5:148691726 | Pro/Ser | 327/769 | Probably damaging |
| ANKRD30A | chr10:37486235 | Glu/Lys | 825/1342 | Possibly damaging |
| BICC1 | chr10:60560788 | Thr/Met | 666/975 | Probably damaging |
| C10orf71 | chr10:50531906 | Pro/Leu | 439/1436 | Probably damaging |
| C1orf96 | chr1:229461083 | Ala/Pro | 238/271 | Probably damaging |
| FAM13C | chr10:61022341 | Leu/Phe | 280/503 | Benign |
| KIAA1731 | chr11:93456335 | Val/Leu | 2026/2602 | Benign |
| OOEP | chr6:74079072 | Thr/Met | 76/150 | Benign |
| OR4K13 | chr14:20502538 | Cys/Tyr | 127/305 | Probably damaging |
| RFX7 | chr15:56385955 | Gly/Asp | 1324/1461 | Benign |
| SECISBP2L | chr15:49304946 | Cys/Arg | 499/1057 | Benign |
AA, amino acid; bp, basepair.
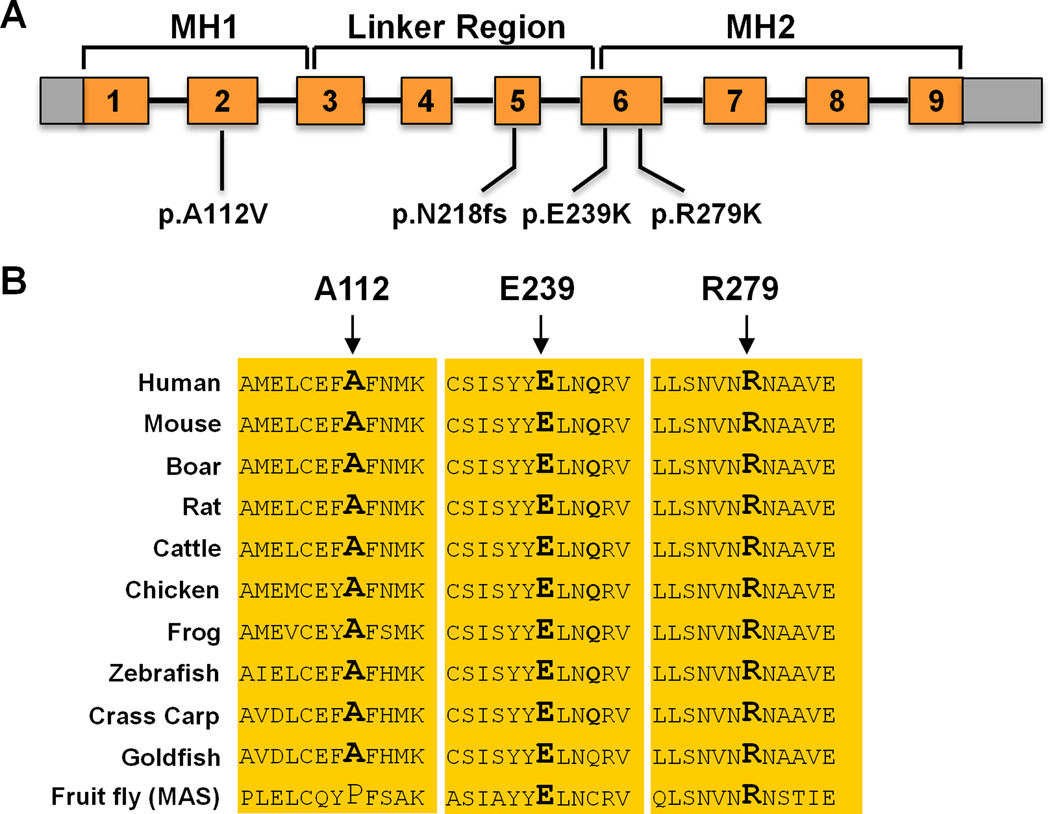
SMAD3 exons and flanking introns were amplified and sequenced as a candidate gene for FTAAD using DNA from 181 unrelated FTAAD probands. Sequencing identified 3 additional variants in four families of European descent: p.R279K (c.836G>A, exon 6) in families TAA071 and TAA072, p.E239K (c.715G>A, exon 6) in family TAA365, and p.A112V (c.235C>T, exon 2) in TAA115. The variants p.R279K and p.E239K alter completely conserved amino acids from human to fruit fly (Figure 2) and are predicted to disrupt protein function based on PolyPhen-2 analysis and HOPE analyses.19 Both mutations are located in the MH2 domain and are predicted to affect hydrogen-bond formations. Furthermore, p.R279K is located within a region that interacts with XPO4 (exportin-4) to promote SMAD3 nuclear export.20 SMAD3 p.A112V is conserved from human to goldfish but not in fruit fly, is predicted to be possibly damaging, and segregates with disease in the family with decreased penetrance. It is notable that clinical features of the SMAD3 p.A112V mutation carriers overlapped with AOS, including thoracic aneurysms involving the aortic root, bifid uvula, scoliosis and early-onset osteoarthritis. Therefore, we have classified this variant as disease-causing but cannot exclude the possibility that it is a benign variant. All variants were not present in approximately 2300 control exomes. In fact, only 5 rare variants were identified in SMAD3 in the 2300 exomes with four of these variants predicted to be benign. Only one variant (p.Y297S) was predicted to disrupt protein function. Segregation of the variants with thoracic aortic disease was confirmed in TAA071, TAA072, TAA365, and TAA115 but decreased penetrance, especially in younger family members, was evident. The SMAD3 mutations segregated in these families with a combined LOD score of 2.69, making the combined LOD score for all families 5.21.
Figure 2. Location and conservation of SMAD3 mutations.
(A) Schematic of the SMAD3 gene shows the location of novel SMAD3 variants identified in FTAAD families. The p.A112V alteration was identified in exon 2 encoding the MH1 domain involved in DNA binding. A frame shift mutation (p.N218fs) was identified in exon 5 leading to premature termination of protein translation and likely to nonsense mediated decay of the RNA. Two additional mutations, p.E239K and p.R279K, were found in exons encoding the MH2 protein-protein binding domain. (B) Orthologue conservation of the SMAD3 protein sequences involving and surrounding the missense mutations identified in the MH1 and MH2 domains.
Phenotypic features associated with the FTAAD in individuals with SMAD3 mutations were compared with the phenotype reported in AOS patients. There are 31 individuals who carry SMAD3 mutations and 11 obligate carriers and family members at 50% risk of inheriting the mutation and presented with aortic dissection. Of these 42 individuals, 21 individuals presented with a thoracic aortic aneurysm and/or dissection, 4 with intracranial aneurysm or subarachnoid hemorrhage, 2 with abdominal aortic aneurysm, and 2 with bilateral iliac aneurysms. The average age at presentation of disease was 45.1 years old (42 years old for thoracic aortic dissections and 51 years old for ICAs or subarachnoid hemorrhage). Of note, two individuals from the TAA071 family who carried the SMAD3 mutation had more than one vascular disease: one member presented with an aortic root aneurysm that was successfully repaired at age of 53 years and then subsequently died at age of 57 years from a subarachnoid hemorrhage, and another member had a Type A dissection at age 37 years and was found to have ectasia of the iliac arteries. Tortuous arteries were noted in 1 out of 17 patients who underwent aortic CT or MR imaging and 2 out of 6 who had similar cerebrovascular imaging. All three individuals belong to family TAA071. Three individuals were noted to have mitral valve prolapse and two individuals had myxomatous mitral valve. One mutation carrier had an atrial septal defect. Two individuals had mild to moderate concentric ventricular hypertrophy. None of the individuals were reported to have other heart defects that were reported in AOS patients, specifically patent ductus arteriosus and atrial fibrillation.
In this cohort of patients, 13 individuals presented with a type A dissection (Stanford classification and defined as dissections initiating in the ascending aorta), one individual with a type B dissection (initiating in the descending aorta just distal to the take off of the subclavian artery), and one individual with an unspecified thoracic aortic dissection. One individual died suddenly at the age of 55 years of unknown cause. The average age of onset of dissection was 42 years (25 – 54 years). There were 8 individuals who had either mild dilatation or aortic root aneurysm (defined as aneurysms at the level of the sinuses of Valsalva) with an average age of 36.6 years at diagnosis (17 – 74 years). Ascending aortic diameters at the time of type A dissection were not reported for most of these individuals, however one individual was noted to have a 50 mm aortic root aneurysm on visual exam at the time of repair of a type A dissection. Three members of the TAA071 had elective repair of their aortic root at 45, 56, and 65 mm, and a member of the TAA072 family is currently 76 years old and has a 47 mm aortic root aneurysm on echocardiographic imaging. It is notable that 13 women with SMAD3 mutations had an average of 3 pregnancies without vascular complications during pregnancy or postpartum.
Cardiovascular and joint disease in SMAD3 mutation carriers are summarized in Table 2. Eight individuals who were evaluated by a geneticist were assessed for phenotypic features of MFS, LDS, and AOS (Figure 1, Online Table I). All families except TAA365 (p.E239K) had at least one member with osteoarthritis. Seven individuals were noted to have osteoarthritis, 2 with degenerative disc or joint disease, and 2 others reported painful joints. Three individuals with osteoarthritis reported age of onset of the disease in their 40s. Those individuals with osteoarthritis reported disease affecting multiple joints, some with deforming or disabling disease. Fourteen out of 25 mutation carriers did not have joint pain or other joint complaints.
Table 2.
Cardiovascular features and joint disease in individuals with SMAD3 mutations.
| Clinical feature | # of patients/total patients assessed |
|---|---|
| Cardiovascular | |
| Thoracic aortic aneurysm/dissection | 21/42 |
| Abdominal aortic aneurysm | 2/42 |
| Iliac artery aneurysms | 2/42 |
| Intracranial aneurysm or subarachnoid hemorrhage | 4/42 |
| Arterial tortuosity (aorta) | 1/17 |
| Arterial tortuosity (cerebral vessels) | 2/6 |
| Mitral valve prolapse | 4/23 |
| Joint disease | |
| Osteoarthritis | 7/25 |
| Degenerative disc or joint disease | 2/25 |
Discussion
Exome sequencing of two distantly affected relatives efficiently and successfully identified a frame shift mutation in SMAD3 as the cause of vascular disease in a family with arterial aneurysms and dissections inherited in an autosomal dominant pattern. Family members with the SMAD3 frame shift mutation presented with ascending thoracic aortic aneurysms leading to aortic dissections and ICAs with subarachnoid hemorrhage, along with abdominal aortic and bilateral iliac artery aneurysms. Subsequent sequencing of families with multiple members with thoracic aortic aneurysms and acute aortic dissections identified SMAD3 mutations in 2% of FTAAD, similar to the frequency of TGFBR2 mutations in FTAAD.8 The vascular disease presentations in these families were primarily TAAD, ICAs and AAAs, and two individuals presenting with TAAD subsequently were diagnosed with vascular disease elsewhere (subarachnoid hemorrhage and iliac ectasia). We had recently described families with autosomal dominant inheritance of TAADs, ICAs, and AAAs, and this study confirms that families with autosomal dominant inheritance of aneurysms in different vascular beds can result from a single gene mutation.5 It is notable that although SMAD3 mutations were identified in some families with TAAD, ICA and AAA, other families described with autosomal dominant inheritance of TAAD/ICA or TAAD/ICA/AAA were not found to have SMAD3 mutations. Therefore, there are additional genes to be identified for the phenotype of presentation with TAAD/ICA/AAA and TAAD/ICA.
SMAD3 mutations were previously reported in families with AOS, a syndrome characterized by aneurysms, dissections and arterial tortuosity, along with early-onset osteoarthritis. The most clinically significant difference between the families described here and the previously reported AOS families was the presentation of ICAs in affected members of FTAAD families. These data suggest that the cerebrovascular circulation needs to be imaged for aneurysms in SMAD3 mutation carriers, in addition to the entire aorta and its branches. These recommendations are similar to the recommendation for TGFBR1 and TGFBR2 mutation carriers.7, 21 In distinct contrast to TGFBR2 mutations, dissections with little to minimal enlargement of the aortic root were not observed in SMAD3 patients. Although some of the SMAD3 mutation carriers in our families had osteoarthritis, which was crippling in a few individuals, the majority of SMAD3 mutation carriers did not report osteoarthritis or joint pain or deformities. However, it is important to note that the AOS families had thorough imaging for osteoarthritis whereas individuals in this study did not undergo imaging and the diagnosis of osteoarthritis and joint pain were based on review of medical records and interviews to assess joint pain. In addition, arterial tortuosity was noted only in a minority of affected members of the FTAAD families despite CT and MRI imaging. Families in this study were recruited worldwide and only a few affected individuals were fully assessed for cutaneous and skeletal features of MFS, LDS and AOS (Online Table I). Although broad or bifid uvula, prominent cutaneous veins, easy bruising, and joint hyperflexibility were noted in a few, these findings were not present in a majority of mutation carriers who were examined.
The SMAD3 mutations identified in FTAAD were found in exons 2, 5 and 6. Previously reported SMAD3 mutations in AOS patients all fell in exon 6. Frame shift mutations were identified in both studies. Although fibroblasts were not available from an individual reported here with the SMAD3 frame shift mutation in exon 5, van de Laar et al. did show that the frame shift mutation in exon 6 results in nonsense mediated decay, suggesting that the frame shift mutation leads to SMAD3 haploinsufficiency. TGF-β initiates cell signaling by binding to its type I and type II receptors (encoded by TGFBR1 and TGFBR2, respectively), inducing phosphorylation and activation of the type I receptor by the type II receptor. The type I receptor kinase then phosphorylates cytoplasmic substrates, including SMAD2 and SMAD3. SMAD2 and SMAD3 are structurally similar but clearly have functionally distinct roles in cell signaling that is perhaps best illustrated by Smad-specific deficient mice. Smad2 null mice die before birth whereas Smad3 null mice live into adulthood.22, 23 Assessment of TGF-β signaling in lung fibroblasts and hepatic stellate cells, which differentiate into myofibroblast with hepatic injury, have suggested that the pathway for TGF-β-driven differentiation fibroblasts and hepatic stellate cells into myofibroblasts involves primarily SMAD3 rather than SMAD2.24, 25 The lack of differentiation of myofibroblasts of Smad3−/− fibroblasts is marked by decreased expression of α-actin, a major marker for differentiated myofibroblasts. The defect in myofibroblast differentiation is illustrated by the finding that Smad3−/− mice are resistant to fibrosis when compared to wildtype mice, including lung fibrosis induced by bleomycin treatment and liver fibrosis induced by dimethylnitrosamine. Studies of cardiac fibroblasts isolated from Smad3−/− mice also showed decreased α-actin expression compared with wild type fibroblasts, along with decreased contraction in collagen gels and decreased production of type III collagen and connective tissue growth factor (CTGF).26 These findings are remarkably similar to analyses of TGFBR2 mutant fibroblasts and smooth muscle cells (SMCs) compared to wild type cells.27 TGFBR2 mutant fibroblasts and SMCs show decreased expression of α-actin, along with other contractile proteins. Published studies of diseased aortic tissue obtained from patients with SMAD3 mutations paradoxically showed increased staining for CTGF and collagen, along with increased phosphorylated SMAD2 and increased SMAD3 protein.12 Further studies are needed to determine how SMAD3 mutations disrupt TGF-β signaling, with careful analysis of TGF-β signaling both at the cellular level and in the diseased aortic tissue. Differences between these systems may result from the fact that the diseased aorta is exposed to altered cell signaling pathways from the progressive disease processes (e.g., cytokines and proteases) and continuous biomechanical forces from the pulsatile blood flow.
In summary, SMAD3 mutations are responsible for 2% of familial thoracic aortic aneurysms and dissections. Mutations are found in families with TAAD alone, along with families with inheritance of a combined phenotype of TAAD, ICAs, and AAAs. Although osteoarthritis, arterial tortuosity, and cutaneous and skeletal features of the MFS, LDS and AOS can be identified in some mutation carriers, these findings are notably absent for many patients from this cohort. SMAD3 mutation should be considered as a possible cause of FTAAD, in particular in those families with features of AOS, families with aortic root aneurysms, or families with ICAs and AAAs in addition to TAAD. Finally, the presentation of aneurysms involving various different arteries in family members harboring the same SMAD3 mutation indicates that these mutations predispose to a diffuse vasculopathy extending beyond the ascending aorta.
Supplementary Material
Novelty and Significance.
What is known?
Thoracic aortic aneurysms leading to acute aortic dissections (TAAD) can be inherited in families in an autosomal dominant pattern with reduced penetrance and variable expression and is designated as familial TAAD.
As part of the spectrum of clinical heterogeneity of familial TAAD, we recently described families with multiple members that presented with TAAD, intracranial aneurysms and abdominal aortic aneurysms inherited in an autosomal dominant manner.
What new information does this article contribute?
An exome sequencing strategy successfully identified a mutation in SMAD3 as the cause of disease in a large family with affected family members presenting with TAAD, intracranial aneurysms and abdominal aortic aneurysms.
The identification of SMAD3 mutations in families with autosomal dominant inheritance of aneurysms involving different vascular beds demonstrates that a single gene mutation can cause a diffuse vasculopathy extending beyond the thoracic ascending aorta.
This study describes the successful and efficient identification of a SMAD3 mutation causing familial TAAD through exome sequencing of DNA from pairs of distantly affected relatives. Subsequent sequencing of unrelated index cases with a family history of TAAD identified SMAD3 mutations in 2% of these families. The vascular disease presentations in these families were primarily TAAD, but affected family members also presented with intracranial aneurysms, abdominal aortic aneurysms, and bilateral ectasia of the iliac arteries. The identification of SMAD3 mutations furthers our understanding of the genes involved in aortic and vascular disease development. Furthermore, these findings suggest that SMAD3 testing should be considered in patients and families with similar disease presentation to allow for early identification of at risk family members and initiation of medical and surgical management in a timely manner to prevent aortic dissections and ruptures and subarachnoid hemorrhages.
Acknowledgments
The authors are extremely grateful to patients involved in this study, and the genetic counselors who aided in the collection of clinical data from the families. We would like to thank the NHLBI GO Exome Sequencing Project and its ongoing studies that produced and provided exome variant calls for comparison: the Lung Cohorts Sequencing Project (HL-102923), the WHI Sequencing Project (HL-102924), the Heart Cohorts Sequencing Project (HL-103010), the Broad Institute Sequencing Project (HL-102925), the Northwest Genomics Center Sequencing Project (HL-102926, D.A.N, M.J.R, and J.S.) and the Family Studies Project Team.
Sources of Funding
The following sources provided funding for the clinical characterization and confirmation sequencing: P50HL083794-01 (D.M.M.), RO1 HL62594 (D.M.M.), UL1 RR024148 (CTSA), the Richard T. Pisani Fund, the Vivian L. Smith Foundation (D.M.M.), and the Doris Duke Distinguished Clinical Scientist Award (D.M.M.).
Non-standard Abbreviations and Acronyms
- TAAD
thoracic aortic aneurysm and/or dissection
- FTAAD
familial TAAD
- ICA
intracranial aneurysm
- AAA
abdominal aortic aneurysm
- MFS
Marfan syndrome
- LDS
Loeys Dietz syndrome
- FBN1
gene for fibrillin 1
- AOS
Aneurysms osteoarthritis syndrome
- TGFBR1
gene for transforming growth factor-β receptor type 1
- TGFBR2
gene for transforming growth factor-β receptor type 2
- ACTA2
gene for smooth muscle aortic α-actin
- bp
base pair
- Indel
genomic insertion or deletion
- del
deletion
- fs
frame shift
- SNVs
single nucleotide variants
- BWA
Burrows-Wheeler Aligner
- PCR
polymerase chain reaction
- LOD
logarithm of the odds
- CTGF
connective tissue growth factor
- SMC
smooth muscle cells
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures
None.
References
- 1.Milewicz DM, Chen H, Park ES, Petty EM, Zaghi H, Shashidhar G, Willing M, Patel V. Reduced penetrance and variable expressivity of familial thoracic aortic aneurysms/dissections. Am J Cardiol. 1998;82:474–479. doi: 10.1016/s0002-9149(98)00364-6. [DOI] [PubMed] [Google Scholar]
- 2.Zhu L, Vranckx R, Khau Van KP, Lalande A, Boisset N, Mathieu F, Wegman M, Glancy L, Gasc JM, Brunotte F, Bruneval P, Wolf JE, Michel JB, Jeunemaitre X. Mutations in myosin heavy chain 11 cause a syndrome associating thoracic aortic aneurysm/aortic dissection and patent ductus arteriosus. Nat Genet. 2006;38:343–349. doi: 10.1038/ng1721. [DOI] [PubMed] [Google Scholar]
- 3.Loscalzo ML, Goh DL, Loeys B, Kent KC, Spevak PJ, Dietz HC. Familial thoracic aortic dilation and bicommissural aortic valve: a prospective analysis of natural history and inheritance. Am J Med Genet A. 2007;143:1960–1967. doi: 10.1002/ajmg.a.31872. [DOI] [PubMed] [Google Scholar]
- 4.Guo DC, Pannu H, Papke CL, Yu RK, Avidan N, Bourgeois S, Estrera AL, Safi HJ, Sparks E, Amor D, Ades L, McConnell V, Willoughby CE, Abuelo D, Willing M, Lewis RA, Kim DH, Scherer S, Tung PP, Ahn C, Buja LM, Raman CS, Shete S, Milewicz DM. Mutations in smooth muscle alpha-actin (ACTA2) lead to thoracic aortic aneurysms and dissections. Nat Genet. 2007;39:1488–1493. doi: 10.1038/ng.2007.6. [DOI] [PubMed] [Google Scholar]
- 5.Regalado E, Medrek S, Tran-Fadulu V, Guo D-C, Pannu H, Golabbakhsh H, Smart S, Chen JH, Shete S, Kim DH, Stern R, Braverman AC, Milewicz DM. Autosomal dominant inheritance of a predisposition to thoracic aortic aneurysms and dissections and intracranial saccular aneurysms. Am J Med Genet A. 2011 doi: 10.1002/ajmg.a.34050. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collod-Beroud G, Le Bourdelles S, Ades L, Ala-Kokko L, Booms P, Boxer M, Child A, Comeglio P, De Paepe A, Hyland JC, Holman K, Kaitila I, Loeys B, Matyas G, Nuytinck L, Peltonen L, Rantamaki T, Robinson P, Steinmann B, Junien C, Beroud C, Boileau C. Update of the UMD-FBN1 mutation database and creation of an FBN1 polymorphism database. Hum Mutat. 2003;22:199–208. doi: 10.1002/humu.10249. [DOI] [PubMed] [Google Scholar]
- 7.Loeys BL, Schwarze U, Holm T, Callewaert BL, Thomas GH, Pannu H, De Backer JF, Oswald GL, Symoens S, Manouvrier S, Roberts AE, Faravelli F, Greco MA, Pyeritz RE, Milewicz DM, Coucke PJ, Cameron DE, Braverman AC, Byers PH, De Paepe AM, Dietz HC. Aneurysm syndromes caused by mutations in the TGF-beta receptor. N Engl J Med. 2006;355:788–798. doi: 10.1056/NEJMoa055695. [DOI] [PubMed] [Google Scholar]
- 8.Pannu H, Fadulu V, Chang J, Lafont A, Hasham SN, Sparks E, Giampietro PF, Zaleski C, Estrera AL, Safi HJ, Shete S, Willing MC, Raman CS, Milewicz DM. Mutations in transforming growth factor-beta receptor type II cause familial thoracic aortic aneurysms and dissections. Circulation. 2005;112:513–520. doi: 10.1161/CIRCULATIONAHA.105.537340. [DOI] [PubMed] [Google Scholar]
- 9.Tran-Fadulu V, Pannu H, Kim DH, Vick GW, III, Lonsford CM, Lafont AL, Boccalandro C, Smart S, Peterson KL, Hain JZ, Willing MC, Coselli JS, LeMaire SA, Ahn C, Byers PH, Milewicz DM. Analysis of multigenerational families with thoracic aortic aneurysms and dissections due to TGFBR1 or TGFBR2 mutations. J Med Genet. 2009;46:607–613. doi: 10.1136/jmg.2008.062844. [DOI] [PubMed] [Google Scholar]
- 10.Milewicz DM, Michael K, Fisher N, Coselli JS, Markello T, Biddinger A. Fibrillin-1 (FBN1) mutations in patients with thoracic aortic aneurysms. Circulation. 1996;94:2708–2711. doi: 10.1161/01.cir.94.11.2708. [DOI] [PubMed] [Google Scholar]
- 11.Francke U, Berg MA, Tynan K, Brenn T, Liu WG, Aoyama T, Gasner C, Miller DC, Furthmayr H. A Gly1127Ser Mutation in An Egf-Like Domain of the Fibrillin-1 Gene Is A Risk Factor for Ascending Aortic-Aneurysm and Dissection. American Journal of Human Genetics. 1995;56:1287–1296. [PMC free article] [PubMed] [Google Scholar]
- 12.van de Laar IM, Oldenburg RA, Pals G, Roos-Hesselink JW, de Graaf BM, Verhagen JM, Hoedemaekers YM, Willemsen R, Severijnen LA, Venselaar H, Vriend G, Pattynama PM, Collee M, Majoor-Krakauer D, Poldermans D, Frohn-Mulder IM, Micha D, Timmermans J, Hilhorst-Hofstee Y, Bierma-Zeinstra SM, Willems PJ, Kros JM, Oei EH, Oostra BA, Wessels MW, Bertoli-Avella AM. Mutations in SMAD3 cause a syndromic form of aortic aneurysms and dissections with early-onset osteoarthritis. Nat Genet. 2011;43:121–126. doi: 10.1038/ng.744. [DOI] [PubMed] [Google Scholar]
- 13.O'Roak BJ, Deriziotis P, Lee C, Vives L, Schwartz JJ, Girirajan S, Karakoc E, MacKenzie AP, Ng SB, Baker C, Rieder MJ, Nickerson DA, Bernier R, Fisher SE, Shendure J, Eichler EE. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat Genet. 2011;43:585–589. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.NHLBI Exome Sequencing Project (ESP) Exome Variant Server. [Accessed on May 2011]; Available at: http://snp.gs.washington.edu/popgenSNP/ [Google Scholar]
- 17.Guo D, Hasham S, Kuang SQ, Vaughan CJ, Boerwinkle E, Chen H, Abuelo D, Dietz HC, Basson CT, Shete SS, Milewicz DM. Familial thoracic aortic aneurysms and dissections: genetic heterogeneity with a major locus mapping to 5q13-14. Circulation. 2001;103:2461–2468. doi: 10.1161/01.cir.103.20.2461. [DOI] [PubMed] [Google Scholar]
- 18.Cottingham RW, Jr, Idury RM, Schaffer AA. Faster sequential genetic linkage computations. Am J Hum Genet. 1993;53:252–263. [PMC free article] [PubMed] [Google Scholar]
- 19.Venselaar H, Te Beek TA, Kuipers RK, Hekkelman ML, Vriend G. Protein structure analysis of mutations causing inheritable diseases. An e-Science approach with life scientist friendly interfaces. BMC Bioinformatics. 2010;11:548. doi: 10.1186/1471-2105-11-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurisaki A, Kurisaki K, Kowanetz M, Sugino H, Yoneda Y, Heldin CH, Moustakas A. The mechanism of nuclear export of Smad3 involves exportin 4 and Ran. Mol Cell Biol. 2006;26:1318–1332. doi: 10.1128/MCB.26.4.1318-1332.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hiratzka LF, Bakris GL, Beckman JA, Bersin RM, Carr VF, Casey DE, Jr, Eagle KA, Hermann LK, Isselbacher EM, Kazerooni EA, Kouchoukos NT, Lytle BW, Milewicz DM, Reich DL, Sen S, Shinn JA, Svensson LG, Williams DM. 2010 ACCF/AHA/AATS/ACR/ASA/SCA/SCAI/SIR/STS/SVM guidelines for the diagnosis and management of patients with Thoracic Aortic Disease: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines, American Association for Thoracic Surgery, American College of Radiology, American Stroke Association, Society of Cardiovascular Anesthesiologists, Society for Cardiovascular Angiography and Interventions, Society of Interventional Radiology, Society of Thoracic Surgeons, and Society for Vascular Medicine. Circulation. 2010;121:e266–e369. doi: 10.1161/CIR.0b013e3181d4739e. [DOI] [PubMed] [Google Scholar]
- 22.Hamamoto T, Beppu H, Okada H, Kawabata M, Kitamura T, Miyazono K, Kato M. Compound disruption of smad2 accelerates malignant progression of intestinal tumors in apc knockout mice. Cancer Res. 2002;62:5955–5961. [PubMed] [Google Scholar]
- 23.Datto MB, Frederick JP, Pan L, Borton AJ, Zhuang Y, Wang XF. Targeted disruption of Smad3 reveals an essential role in transforming growth factor beta-mediated signal transduction. Mol Cell Biol. 1999;19:2495–2504. doi: 10.1128/mcb.19.4.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uemura M, Swenson ES, Gaca MD, Giordano FJ, Reiss M, Wells RG. Smad2 and Smad3 play different roles in rat hepatic stellate cell function and alpha-smooth muscle actin organization. Mol Biol Cell. 2005;16:4214–4224. doi: 10.1091/mbc.E05-02-0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gu L, Zhu YJ, Yang X, Guo ZJ, Xu WB, Tian XL. Effect of TGF-beta/Smad signaling pathway on lung myofibroblast differentiation. Acta Pharmacol Sin. 2007;28:382–391. doi: 10.1111/j.1745-7254.2007.00468.x. [DOI] [PubMed] [Google Scholar]
- 26.Dobaczewski M, Bujak M, Li N, Gonzalez-Quesada C, Mendoza LH, Wang XF, Frangogiannis NG. Smad3 signaling critically regulates fibroblast phenotype and function in healing myocardial infarction. Circ Res. 2010;107:418–428. doi: 10.1161/CIRCRESAHA.109.216101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Inamoto S, Kwartler CS, Lafont AL, Liang YY, Fadulu VT, Duraisamy S, Willing M, Estrera A, Safi H, Hannibal MC, Carey J, Wiktorowicz J, Tan FK, Feng XH, Pannu H, Milewicz DM. TGFBR2 Mutations Alter Smooth Muscle Cell Phenotype and Predispose to Thoracic Aortic Aneurysms and Dissections. Cardiovasc Res. 2010;88:520–529. doi: 10.1093/cvr/cvq230. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.