Abstract
MicroRNAs (miRNAs) are important regulators of eukaryotic gene expression in most biological processes. They act by guiding the RNAi-induced silencing complex (RISC) to partially complementary sequences in target mRNAs to suppress gene expression by a combination of translation inhibition and mRNA decay. The commonly accepted mechanism of miRNA targeting in animals involves an interaction between the 5′-end of the miRNA called the ‘seed region’ and the 3′ untranslated region (3′-UTR) of the mRNA. Many target prediction algorithms are based around such a model, though increasing evidence demonstrates that targeting can also be mediated through sites other than the 3′-UTR and that seed region base pairing is not always required. The power and validity of such in silico data can be therefore hindered by the simplified rules used to represent targeting interactions. Experimentation is essential to identify genuine miRNA targets, however many experimental modalities exist and their limitations need to be understood. This review summarizes and critiques the existing experimental techniques for miRNA target identification.
INTRODUCTION
MicroRNAs (miRNAs) control gene expression post-transcriptionally by binding to complementary sequences in target mRNAs, thereby guiding the effector proteins of RNAi-induced silencing complex (RISC) into close proximity with the mRNA. Complete complementarity between miRNA:mRNA pairs is rare in mammals, but as little as a 6 bp match with the target mRNA can be sufficient to suppress gene expression (1–4). With the exception of a handful of miRNAs reported to increase expression of a target gene (5,6), miRNAs repress gene expression by a combination of mRNA degradation and translation inhibition. They can promote mRNA degradation by either of two mechanisms: direct Argonaute2-catalysed endonucleolytic cleavage of the target (7–10), or deadenylation and exonucleolytic attack, which is the predominant mechanism for miRNA activity in mammals (11). Direct cleavage by Argonaute2 only occurs when there is near perfect complementarity between the miRNA and target mRNA (12), a situation that occurs much more frequently in plants (13) than in mammals (10,14,15). For a detailed review of miRNA mechanisms of actions see Krol et al. (16).
The mechanism by which miRNA sequence complementarity conveys functional binding to mRNA targets has been studied at length, providing rules for miRNA target prediction algorithms. One commonly accepted rule is that the 5′ region of a miRNA from nucleotides 2 to 8 (known as the ‘seed’ region) has particular importance in targeting, as demonstrated by numerous biochemical and structural findings (17). The seed region is the most evolutionarily conserved region of miRNAs (1,18), it is the region that is most frequently complementary to target sites in 3′ untranslated region (3′-UTRs) (19) and in many instances a seed match alone is sufficient to confer mRNA recognition (1,2,20,21). Despite the importance of the seed region, the 3′-end of a miRNA also contributes to effective binding in roughly 2% of all preferentially conserved sites (22,23). Furthermore, some validated miRNA target sites do not have a complete seed match but instead exhibit 11–12 continuous base pairs in the central region of the miRNA (15). As new experimental data like this come to hand the accepted modes of miRNA targeting are expanded, although prediction programs may not incorporate all these experimentally derived possibilities. For example the three most commonly used bioinformatic target prediction tools Targetscan, miRanda and PicTar search for miRNA targets exclusively in mRNA 3′-UTRs and do not incorporate evidence of functional targeting within the 5′-UTR and protein coding region (22,24–26). Also, most algorithms do not adjust predictions for the co-expression of miRNA and target which is proposed to be an effective way of improving predictions (27).
The various miRNA target prediction programs, which use different rules of targeting, produce rather different lists of predicted targets. Differences can arise from the source of 3′-UTR sequences; Targetscan uses the Ensembl database to define 3′-UTRs (28), whereas miRanda uses the University of California Santa Cruz (UCSC) database (29). This alone manifests in major differences between prediction outcomes. Nevertheless, if the Targetscan algorithm is applied to the two separate 3′-UTR databases only a 47% overlap of predicted targets is observed. Similarly if the miRanda algorithm is applied to both 3′-UTR databases separately only a 65% overlap is observed (27). With the identification of genuine miRNA targets lacking a complete 6-mer match (15,30–32) and the further complications of RNA structure and RNA-binding proteins affecting site accessibility (33,34), many predictions may not be bona fide targets and many genuine targets can be missed (35). Accordingly, the false positive rate of prediction programs has been variously calculated to be 24–70% (36–40). This underscores the requirement for experimental data to demonstrate genuine miRNA targets and miRNA function.
GENE-SPECIFIC EXPERIMENTAL VALIDATION OF miRNA TARGETS
Gene-specific experimental validation with the well-established techniques of qRT–PCR, luciferase reporter assays and western blot are commonly used to indicate individual miRNA:mRNA interactions. For a detailed review of methods for the experimental validation of specific miRNA targets refer to Kuhn et al. (41). Generally, the downstream effects of differential miRNA expression are observed at the protein level by western blot and at the mRNA level by qRT–PCR, although these measures will not distinguish between direct and secondary miRNA targets. Reporter assays have been employed extensively to demonstrate a direct link whereby expression of a luciferase reporter—3′-UTR construct will be altered through manipulation of a regulatory miRNA. Direct miRNA effects are demonstrated by the loss of regulation in constructs with mutated miRNA target sites. The disadvantages of reporter assays are that they are labour intensive, dependent upon the region chosen for cloning and can be sensitive to variances in protocol such as the method of transfection or promoter identity (42–44).
In the specific circumstance where a miRNA target is directly cleaved, RNA ligase mediated—5′ rapid identification of cDNA ends (5′ RLM-RACE) may be used to confirm such targeting. Briefly 5′ RLM-RACE is a PCR-based technique, whereby an RNA adapter is ligated to the free 5′ phosphate of an uncapped mRNA produced from, among other nucleolytic activities, Argonaute2-directed mRNA cleavage. The ligation product can be reverse transcribed using a forward primer directed against the linker and a gene specific reverse primer which is subsequently PCR amplified, cloned and identified by sequencing. 5′ RLM-RACE has been used to support direct cleavage of HOXB8 by miR-196 in the mouse embryo (45), validate parallel analysis of RNA ends (PARE) results in mammals (10,14,15), and has been employed extensively to validate products of RISC-mediated cleavage in plants (46).
EXPERIMENTAL miRNA TARGET SCREENING TECHNIQUES
Demonstrating individual miRNA:mRNA interactions misses the capacity for miRNAs to regulate complex gene networks. Uncovering networks requires large scale and unbiased methods of miRNA target identification. To date, the majority of large-scale miRNA target identification experiments involve differential expression of a single miRNA followed by downstream gene-expression or proteomic analysis. Most commonly, differential expression is attained by exogenous expression of a miRNA, however inhibition of an endogenous miRNA is also possible. Over-expression can be achieved by transient transfection of a synthetic miRNA precursor or by stable introduction, typically with a lentiviral vector, of a miRNA expression construct. miRNA inhibition can be achieved by expressing modified antisense oligonucleotides able to bind mature miRNAs and block their activity. There are a variety of miRNA silencing chemistries including anti-miRs (47,48), antagomiRs (49), miRNA sponges (50) and TuD (tough decoy) constructs (51).
Caveats of miRNA over-expression
Despite widespread use, miRNA over-expression experiments are subject to a degree of scepticism for their potential to generate false positive results brought about through the supraphysiological increase in miRNA levels generally achieved after transient transfection (52). Although much of the transfected pre-miR may not be incorporated into RISC complexes to be functionally active, this may still be considerably greater increase than the 20–30% range by which many endogenous miRNAs fluctuate to modulate gene expression (53). Such exaggerated miRNA over-expression can potentially saturate RISC complexes and displace other endogenous miRNAs (54) and consequently cause low affinity target sites to appear functionally important. The use of miRNA mimics is a common approach to transiently over-express miRNAs, however it bypasses the natural mechanism of miRNA biosynthesis, whereby a transcribed pri-miRNA is processed by Dicer and Drosha to form the miRNA:miRNA* duplex. Instead mimics are typically chemically synthesized duplexes that are designed with the aim of activation of only one miRNA strand. The passenger strand will not necessarily be equivalent to the natural miRNA* form, but the potential still exists for this strand to be incorporated into RISC and mediate off-target effects. Another consideration is that over-expression experiments are commonly performed in a cell environment that is artificial to the chosen miRNA, in which case cell-specific natural targets may be missed, while other targets not normally co-expressed with the miRNA are detected. For example several studies over-express brain specific miR-124 in ovarian cancer (HeLa) cells (24,26,55).
The inherent problems of over-expression may be avoided by using miRNA silencing to achieve differential expression of a miRNA within physiological limits. However, miRNA silencing by antisense oligonucleotides is limited by being only as specific as the inhibitor used. For example, this approach may not be selective enough to distinguish between members of the same miRNA family with similar sequences. Additionally, antisense oligonucleotides may sequester endogenous miRNAs without aiding their decay. This means that miRNA quantitation by qRT–PCR will not measure a decrease in miRNA levels following miRNA inhibition, making it difficult to determine the silencing efficiency. When miRNA inhibition and over-expression strategies are compared, as has been performed with cartilage-specific miR-140, the changes in miRNA abundance after inhibition were not as great as in a separate over-expression model, though the mRNA targets identified are theoretically more biologically relevant (56).
Gene expression analysis
Degradation of target mRNAs following ectopic miRNA expression is observable on a genome-wide scale by microarray analysis. The first reported use of this strategy demonstrated that following the transfection of miR-1 or miR-124 mimics into HeLa cells, more than 100 mRNAs were down-regulated in each case. Supporting direct targeting of many of these genes, a 3′-UTR motif search revealed the 6 nt consensus sequences matching the seed regions of miR-1 (CAUUCC) and miR-124 (GUGCCU) were present in 88 and 76% of down-regulated genes, respectively. When the same experiment was performed with a miR-124 mimic with a mutated seed region, the down-regulated genes were not enriched for miR-124 seed matches (26). Since then, similar results have been observed in multiple cellular contexts with other miRNAs (22,26,57,58). Technical advances in next-generation sequencing technology have now enabled the use of RNA-seq as an alternative to microarray gene expression analysis, allowing a deeper analysis to provide a larger list of inferred miRNA targets in comparable over-expression studies (59).
The limitation of using differential gene expression to identify miRNA targets is that they are observed amongst a pool of indirect changes in transcript abundance. This may assist in describing the predominate genes and pathways affected by a miRNA but does not distinguish between direct targets. Seed matches provide one way of enriching for direct miRNA targets over secondary effects, and bioinformatic tools for mining miRNA targets across large-scale gene expression studies have been developed (60). Nevertheless, identifying direct targets remains problematic given the modest effect on levels of some target mRNAs and the fact some miRNA targeting occurs predominantly at the level of translational repression (26). Despite this, the use of gene expression analysis to find miRNA targets is endorsed by a recent report in which at least 84% of miRNA mediated repression was attributable to decreased mRNA abundance (61).
Immunoprecipitation of RISC components
Biochemical approaches have been developed to aid the identification of direct miRNA targets. miRNA:mRNA target pairs can be purified by the immunoprecipitation of the RISC components, Argonaute (AGO) (24,25,38,57,62,63) or TNRC6 (63). Target mRNAs undergoing direct regulation are co-immunoprecipitated along with RISC and identified by microarray or deep sequencing. Argonaute co-immunoprecipitation has been applied to identify targets of the well studied miRNAs miR-1 and miR-124 (25,57). In these examples an exogenous, epitope-tagged Argonaute (AGO2 or AGO1) was expressed in HeLa or HEK-293 T cells together with miR-1 or miR-124, then immunoprecipitated using an antibody against the epitope tag. Precipitates were analysed by microarray in comparison to a mock sample. Illustrating the success of the technique, mRNAs co-immunoprecipitated with Argonaute from cells with ectopic miRNA expression were enriched for seed sequences, with 70% of the miR-1 targets and 75% of the miR-124 targets having a 6-mer seed sequence. This degree of enrichment was highly significant over what would be expected by chance (25). Similar Argonaute immunoprecipitation methods have been performed in different cell contexts, including using a hemagglutinin (HA)-tagged AGO1 in Drosophila Melanogaster Schneider SL2 (S2) cells (38) and co-immunoprecipitation of endogenous AGO from HEK293 (62), and from mouse cardiac muscle (64). Unlike global gene expression analysis experiments with miRNA over-expression, capturing active miRNA:mRNA target pairs allows identification of mRNAs regulated at both the level of degradation and translational repression.
One potential drawback of the Argonaute co-immunoprecipitation approach is that it may not necessarily reflect in vivo interactions between molecules if interactions between RNA and RNA-binding proteins occur subsequent to cell lysis (65). This would artificially facilitate interactions between RNA and proteins that are usually segregated by cellular compartments. In addition, this methodology relies upon a sufficiently stable interaction between the miRNA–mRNA target and the AGO proteins to survive the co-immunoprecipitation process. While the above reports were clearly successful in target enrichment, the potential for loss of targets during this process is uncertain.
High-throughput sequencing of RNA isolated by crosslinking immunoprecipitation
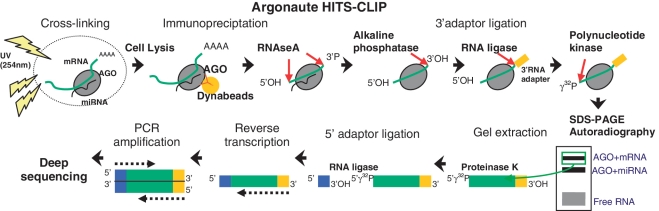
To ensure co-immunoprecipitation reflects cellular interactions, an advancement on the above-mentioned technique utilizes ultraviolet irradiation to crosslink RNA to associated RNA-binding proteins prior to immunoprecipitation, followed by deep sequencing to comprehensively identify bound RNAs (Figure 1). This technique [alternately known as high-throughput sequencing of RNA isolated by crosslinking immunoprecipitation (HITS-CLIP), or crosslinking immunoprecipitation (CLIP)-seq] was first applied to mouse brain (24) and subsequently to Caenorhabditis elegans (66) to identify AGO-bound miRNA:mRNA complexes. These analyses have provided compelling data on the location of miRNA binding sites, within both the 3′-UTR and coding sequence, and generated genome-wide interaction maps for both exogenously expressed miR-124 and general endogenous miRNA targeting (24). Performing HITS-CLIP on cells with and without treatment with specific miRNA antisense inhibitors is likely to provide a powerful method for identification of specific miRNA targets.
Figure 1.
Argonaute high throughput sequencing of cross-linking immunoprecipitation (HITS-CLIP) protocol, also known as CLIP-seq.
HITS-CLIP is a powerful technique capable of providing an extensive insight to the location of miRNA targeting within an mRNA. However, it has been criticized for being limited by the low efficiency of UV 254 nm RNA–protein crosslinking (63). Furthermore HITS-CLIP reads do not precisely pinpoint the position of crosslinking between the RNA and protein (63), and thus can only identify a targeted region (∼100-nt) as opposed to a specific target site. Many sites identified by HITS-CLIP have been validated (24,66), however it is not clear what percentage of clusters represent genuine target sites.
Photoactivatable-ribonucleoside-enhanced crosslinking and immunoprecipitation
A modified crosslinking immunoprecipitation method for isolating protein-associated RNAs, termed photoactivatable-ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP) (63), has been developed to offer more efficient UV crosslinking by incubating cultured cells with a photoactivatable nucleoside such as 4-thiouridine. This improves RNA recovery by 100- to 1000-fold compared to the HITS-CLIP methodology previously described and also is capable of identifying the location of the crosslink and thus more precisely indicate the site of targeting. This is achieved because the 4-thiouridine that incorporates into RNA during co-incubation in cell culture results in thymidine (T) to cytidine (C) transitions more frequently in cross-linked than non-cross-linked sites, thereby marking sites of direct interaction (63).
In one study by Hafner et al. (63), PAR-CLIP was performed using epitope tagged Argonaute family members (AGO 1–4) expressed in HEK293T cells. The most significantly enriched 7-mer motifs identified in co-immunoprecipitated RNA corresponded to the seed sequences of the most abundant miRNAs which were generally positioned 1–2 nt downstream of the predominant crosslinking site. This places the site of crosslinking near the centre of the AGO–miRNA–mRNA complex and illustrates the capacity for using T–C transitions in sequenced DNA to more specifically hone in on miRNA:mRNA interaction sites. Comparable to HITS-CLIP data from the Darnell lab (24), 46% of miRNA binding sites were mapped to 3′-UTRs, 50% to the mRNA coding region and 4% to 5′-UTRs. Target sites were validated by inhibiting the 25 most highly expressed miRNAs using 2′-O-methyl-modified antisense oligoribonucleotides followed by microarray gene expression analysis. mRNAs with miRNA binding sites were more likely to be up-regulated after miRNA silencing, with up-regulation being most frequent when these sites were located within 3′-UTRs.
PAR-CLIP of the TNRC6 family, another RISC component (63), gave reads of which >50% were within 25 nt of Argonaute crosslinked sites, demonstrating mRNA targets are in sufficiently close proximity to both RISC components to undergo UV crosslinking. PAR-CLIP of Argonaute was more efficient than TNRC6, yielding 4000 clusters compared to 600.
Biotin tagged miRNA
In another biochemical approach to enrich for miRNA targets, Orom and Lund (67) transfected cells with biotinylated miRNA duplexes and captured miRNA:mRNA complexes from cell lysates using streptavidin beads. This technique has been applied in both Drosophila and mammalian cell lines to independently demonstrate previously-defined targets of the miRNAs bantam and miR-124a. In the case of miR-124a, the technique enriched the known target LAMC1 roughly 5-fold compared to a control miRNA, and >100-fold compared to mock-transfected cells (68). Microarray analysis from the immunoprecpipitation of biotin tagged miR-10a from mouse E14 embryonic stem cells showed a significant enrichment of mRNAs encoding ribosomal proteins. It was further reported that miR-10a targets ribosomal mRNAs within their 5′-UTR to increase translation (68). A potential advantage of this technique over the immunoprecipitation of RISC components is that in principal it can specifically pull down targets of a single miRNA, although the caveats of miRNA over-expression that we discuss above need be considered. It is not known what affect the biotin tag has on miRNA binding and the ability of this technique to comprehensively identify true miRNA targets has yet to be fully demonstrated.
Detection of direct cleavage targets
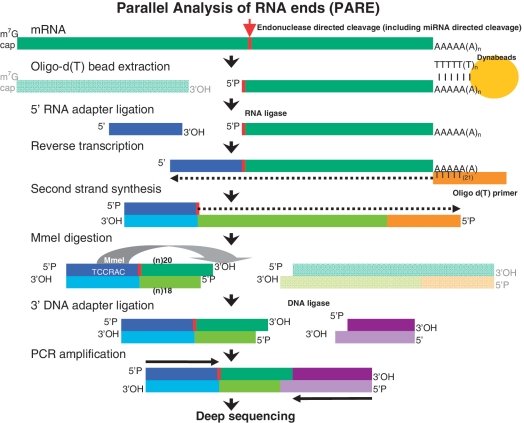
Parallel analysis of RNA ends (PARE) identifies mRNA cleavage products on a global scale by high-throughput sequencing of products from a modified 5′ RNA ligase mediated-rapid amplification of cDNA ends (5′ RLM-RACE) procedure. This takes advantage of the free 5′-monophosphate remaining on the 3′ fragment after Argonaute-mediated cleavage, to which an RNA adaptor can be ligated without additional chemical modification (Figure 2). Subsequent reverse transcription and PCR amplification then enriches these products, prior to deep sequencing and genomic mapping. PARE, also known as degradome-seq or genome-wide mapping of uncapped transcripts (GMUCT) (69,70), has been used to identify widespread mRNA cleavage events regulated by miRNAs in Arabidopsis (13,71), rice (72) and limited cleavages in mammals (10,14,15). Because extensive base pairing between miRNAs and mRNAs leading to direct RISC-mediated cleavage does not appear to be a major mechanism of miRNA activity in mammals, the use of PARE is most suited to plant systems, where it identifies the large subset of miRNA targets that are subject to direct cleavage (73).
Figure 2.
PARE (parallel analysis of RNA ends) protocol, also known as GMUCT (genome-wide mapping of uncapped transcripts) or degradome-seq.
Reverse transcription of targets
In a method for detecting miRNA–mRNA complexes developed by Vatolin and colleagues (74), endogenous miRNAs are used as primers for cDNA synthesis of target mRNAs. Because miRNAs bind their mRNA target within the RISC, a strong detergent is used to disassociate these proteins to allow reverse transcriptase to bind and synthesize cDNA. The cDNA can then be cloned and sequenced to identify the bound mRNA. This method has thus far only been applied to C. elegans, where it successfully identified the well established interaction between the miRNA lin-4 and lin-14 as well as showing a novel interaction between let-7 and its target K10C3.4 (75).
Proteomic approaches
Stable isotope labelling with amino acids in cell culture
Stable isotope labelling with amino acids in cell culture (SILAC) is a high-throughput method for quantitative proteomics in which relative protein abundance is measured by mass spectrometry of samples labelled with different isotopes. Because a significant degree of miRNA activity is mediated at the level of translation (40), proteomic approaches have an inherent advantage of assaying the ultimate effect of miRNAs. SILAC has been previously applied to measure the effect of an over-expressed miRNA on the proteome by comparing miRNA-transfected to mock-transfected cells. For example, SILAC of HeLa cells transfected with miR-1 revealed the repression of 12 proteins (from a set of 504 detected proteins) for which there was a significant enrichment of miR-1 seed sequence sites (55). Proteomic investigations are more limited in their depth of coverage than other gene expression strategies, but as technologies improve this should become less problematic. Indeed, subsequent uses of SILAC have demonstrated that single miRNAs are capable of repressing the production of hundreds of proteins (39,40), directly or indirectly. Using SILAC to identify targets of miR-143, Yang and colleague’s (76) compared miR-143 mimic- to control-transfected MiaPaCa2 pancreatic cancer cells. They identified over 1200 proteins of which 93 were down-regulated more than 2-fold. Luciferase reporter assays of 34 of these showed that 10 were likely direct miR-143 targets.
Two-dimensional differentiation in-gel electrophoresis
Two-dimensional differentiation in-gel electrophoresis (2D-DIGE) profiling is another proteomic approach that has been applied to the identification of miRNA targets. It involves electrophoresis on a single gel of two samples labelled with different fluorescent dyes, separating the proteins by iso-electric focusing and SDS–PAGE and then identifying them by mass spectrometry. 2D-DIGE has been applied to the investigation of miR-21 targets in MCF7 cells treated with anti-miR inhibitor of miR-21 (77). Mass spectrometry revealed seven up-regulated proteins, of which three were validated by western blot, qRT–PCR and reporter assays. In a separate study of cells transfected with either a miR-29a mimic or antisense inhibitor, over 100 differentially regulated proteins were identified, with fluctuations in level generally being modest (between 1.2 and 1.7-fold) (78). Only 14 of these mRNAs contained miR-29a seed sequences in their 3′-UTR, far less than comparable gene expression analysis experiments (26).
Translation profiling
The analysis of mRNAs associated with elongating ribosomes identifies translationally active mRNAs. In the technique of ‘polysome profiling’, cyclohexamide is used to trap elongating ribosomes. Centrifugation through a sucrose gradient then separates mRNAs with no associated ribosomes from those with bound ribosomes which are presumably undergoing translation. The polysome profile of an mRNA provides information on two key parameters of translation; the fraction of the mRNA species bound by at least one ribosome (referred to as ribosome occupancy) and the average number of ribosomes bound per 100 bases of coding sequence (referred to as ribosome density) (44). Poly(A)+ RNAs from bound and unbound pools are isolated, amplified, coupled to Cy5 and Cy3 dyes, respectively, and competitively hybridized to DNA microarrays. Polysome profiling of HEK-293 T cells with and without over-expression of miR-124 has been used to determine the relative contribution of translational repression and mRNA degradation in mediating miRNA activity (44). Translation profiles for ∼8000 genes were obtained, revealing around 600 putative miR-124 targets.
In the related method of ‘ribosome profiling’ or ‘ribosome footprinting’, cells are lysed after cyclohexamide treatment then treated with RNAse I to degrade mRNAs not protected by ribosomes. The resulting 80 S monosomes are purified on sucrose gradients and the protected mRNA fragments identified by high-throughput sequencing. Ribosome profiling has been applied to HeLa cells transfected with either miR-1, miR-155 or miR-223 (61). By comparing the effect upon translation determined through ribosome profiling with changes in mRNA level obtained by microarray analysis, the authors concluded that at least 84% of miRNA-mediated repression was attributable to mRNA degradation (61). Although translational profiling methods do not directly measure protein levels they provide quantative data at a greater depth than currently possible by proteomic approaches, giving a powerful methodology to determine miRNA activity.
FINDING MULTIPLE miRNAs THAT TARGET A SINGLE mRNA
While a single miRNA can target many genes, multiple miRNAs can regulate a single gene (79,80), and methods to comprehensively identify miRNAs that regulate individual genes of interest have been developed. In one approach individual miRNAs are successively transfected into a cell line that stably expresses a luciferase reporter containing the 3′-UTR of the target mRNA of interest. In a study aiming to identify miRNAs targeting p21/Waf, cells expressing the luciferase-3′-UTR reporter gene were individually transfected with 266 miRNA mimics that had been bioinformatically predicted to target p21/Waf1 (79). Of these 266 miRNAs tested, 28 suppressed reporter activity, including all of the miRNAs previously reported to target p21/Waf1. Using a similar method, Jiang and colleagues found that 7 out of 45 miRNAs tested repress a CyclinD1 3′-UTR luciferase reporter (81).
To identify the miRNAs targeting the transcription factor Hand2, Vo and colleagues (80) used an affinity purification method. The 3′-UTR of Hand2 was fused to an MS2 tag and cloned downstream of a GFP reporter, which was then transduced into dissociated rat neonatal cardiomyocytes. Complexes on the chimeric GFP-Hand2-MS2 mRNA were isolated from cell lysates using an affinity column containing bound MS2 binding protein, and the associated miRNAs were identified using multiplex PCR miRNA arrays. One of the identified miRNAs was miR-1, which had previously been found to target Hand2, thereby validating the affinity capture approach. MiR-133a was also identified and its targeting of Hand2 was subsequently verified by mutating miR-133a binding sites within the Hand2 3′-UTR, which abrogated its capture by the affinity column. The targeting of Hand2 by miR-133a was further verified by its effect on Hand2 mRNA and protein levels and in luciferase reporter experiments.
These techniques can demonstrate instances where a single gene is targeted by multiple miRNAs. In contrast to the reporter assay approach, the affinity purification method has the advantage that it is capable of demonstrating direct binding, also it does not require foreknowledge of potential targeting miRNAs and does not rely upon miRNA over-expression. A significant finding was also made possible using the affinity purification protocol that both miR-1 and miR-133a simultaneously bind to the Hand2 3′-UTR and synergistically regulate Hand2 expression. This was tested by co-transfecting biotinylated miR-1 with the MS2-tagged Hand2 3′-UTR followed by consecutive affinity purification with the MS2 binding protein column and streptavidin beads. miR-1 and miR-133a were both identified by qRT–PCR (80). However, both of these techniques are limited to the region of the mRNA chosen for inclusion in the hybrid mRNA. Typically this is the 3′-UTR, which means miRNAs targeting the 5′-UTR or coding region will not be identified.
CONCLUSION
Each individual miRNA is likely to down-regulate the abundance and/or translation of many mRNAs (26,39,40,61). Compounding the complexity of miRNA control, multiple miRNAs can act together on individual mRNAs to produce additive or synergistic effects on protein production (79,80). Thus, miRNA research will increasingly focus upon miRNA-regulated networks (82), in addition to identifying individual miRNA:mRNA interactions. Multiple methodologies are now available to ascertain miRNA targeting, each with intrinsic strengths and weaknesses as discussed above. Combining multiple strategies is required to obtain a comprehensive high-confidence description of miRNA targeting networks.
FUNDING
This work was supported by grants from the National Health & Medical Research Council (NHMRC) (to C.P.B., G.J.G.); the National Breast Cancer Foundation (Research Fellowship to C.P.B.); the University of Adelaide (PhD scholarship to D.W.T). Funding for open access charge: National Health and Medical Research Council and Cancer Research, South Australia.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank members of the Goodall lab for useful discussion.
REFERENCES
- 1.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 2.Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lai EC, Tam B, Rubin GM. Pervasive regulation of Drosophila Notch target genes by GY-box-, Brd-box-, and K-box-class microRNAs. Genes Dev. 2005;19:1067–1080. doi: 10.1101/gad.1291905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc. Natl Acad. Sci. USA. 2008;105:1608–1613. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 7.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 8.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 9.Okamura K, Ishizuka A, Siomi H, Siomi MC. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 2004;18:1655–1666. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bracken CP, Szubert JM, Mercer TR, Dinger ME, Thomson DW, Mattick JS, Michael MZ, Goodall GJ. Global analysis of the mammalian RNA degradome reveals widespread miRNA-dependent and miRNA-independent endonucleolytic cleavage. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr110. doi:10.1093/nar/gkr110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 12.Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.German MA, Pillay M, Jeong DH, Hetawal A, Luo S, Janardhanan P, Kannan V, Rymarquis LA, Nobuta K, German R, et al. Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat. Biotechnol. 2008;26:941–946. doi: 10.1038/nbt1417. [DOI] [PubMed] [Google Scholar]
- 14.Karginov FV, Cheloufi S, Chong MM, Stark A, Smith AD, Hannon GJ. Diverse endonucleolytic cleavage sites in the mammalian transcriptome depend upon microRNAs, Drosha, and additional nucleases. Mol. Cell. 2010;38:781–788. doi: 10.1016/j.molcel.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shin C, Nam JW, Farh KK, Chiang HR, Shkumatava A, Bartel DP. Expanding the microRNA targeting code: functional sites with centered pairing. Mol. Cell. 2010;38:789–802. doi: 10.1016/j.molcel.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 17.Jinek M, Doudna JA. A three-dimensional view of the molecular machinery of RNA interference. Nature. 2009;457:405–412. doi: 10.1038/nature07755. [DOI] [PubMed] [Google Scholar]
- 18.Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, Rhoades MW, Burge CB, Bartel DP. The microRNAs of Caenorhabditis elegans. Genes Dev. 2003;17:991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stark A, Brennecke J, Russell RB, Cohen SM. Identification of drosophila microRNA targets. PLoS Biol. 2003;1:E60. doi: 10.1371/journal.pbio.0000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 21.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 22.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shkumatava A, Stark A, Sive H, Bartel DP. Coherent but overlapping expression of microRNAs and their targets during vertebrate development. Genes Dev. 2009;23:466–481. doi: 10.1101/gad.1745709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hendrickson DG, Hogan DJ, Herschlag D, Ferrell JE, Brown PO. Systematic identification of mRNAs recruited to argonaute 2 by specific microRNAs and corresponding changes in transcript abundance. PLoS ONE. 2008;3:e2126. doi: 10.1371/journal.pone.0002126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 27.Ritchie W, Flamant S, Rasko JE. Predicting microRNA targets and functions: traps for the unwary. Nat. Methods. 2009;6:397–398. doi: 10.1038/nmeth0609-397. [DOI] [PubMed] [Google Scholar]
- 28.Flicek P, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, et al. Ensembl 2008. Nucleic Acids Res. 2008;36:D707–D714. doi: 10.1093/nar/gkm988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bina M. The genome browser at UCSC for locating genes, and much more! Mol. Biotechnol. 2008;38:269–275. doi: 10.1007/s12033-007-9019-2. [DOI] [PubMed] [Google Scholar]
- 30.Didiano D, Hobert O. Perfect seed pairing is not a generally reliable predictor for miRNA-target interactions. Nat. Struct. Mol. Biol. 2006;13:849–851. doi: 10.1038/nsmb1138. [DOI] [PubMed] [Google Scholar]
- 31.Lal A, Navarro F, Maher CA, Maliszewski LE, Yan N, O’Day E, Chowdhury D, Dykxhoorn DM, Tsai P, Hofmann O, et al. miR-24 Inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to “seedless” 3′UTR microRNA recognition elements. Mol. Cell. 2009;35:610–625. doi: 10.1016/j.molcel.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vella MC, Choi EY, Lin SY, Reinert K, Slack FJ. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3'UTR. Genes Dev. 2004;18:132–137. doi: 10.1101/gad.1165404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ameres SL, Martinez J, Schroeder R. Molecular basis for target RNA recognition and cleavage by human RISC. Cell. 2007;130:101–112. doi: 10.1016/j.cell.2007.04.037. [DOI] [PubMed] [Google Scholar]
- 34.Kedde M, Agami R. Interplay between microRNAs and RNA-binding proteins determines developmental processes. Cell Cycle. 2008;7:899–903. doi: 10.4161/cc.7.7.5644. [DOI] [PubMed] [Google Scholar]
- 35.Alexiou P, Maragkakis M, Papadopoulos GL, Reczko M, Hatzigeorgiou AG. Lost in translation: an assessment and perspective for computational microRNA target identification. Bioinformatics. 2009;25:3049–3055. doi: 10.1093/bioinformatics/btp565. [DOI] [PubMed] [Google Scholar]
- 36.Bentwich I. Prediction and validation of microRNAs and their targets. FEBS Lett. 2005;579:5904–5910. doi: 10.1016/j.febslet.2005.09.040. [DOI] [PubMed] [Google Scholar]
- 37.Sethupathy P, Megraw M, Hatzigeorgiou AG. A guide through present computational approaches for the identification of mammalian microRNA targets. Nat. Methods. 2006;3:881–886. doi: 10.1038/nmeth954. [DOI] [PubMed] [Google Scholar]
- 38.Easow G, Teleman AA, Cohen SM. Isolation of microRNA targets by miRNP immunopurification. RNA. 2007;13:1198–1204. doi: 10.1261/rna.563707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 40.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuhn DE, Martin MM, Feldman DS, Terry AV, Jr, Nuovo GJ, Elton TS. Experimental validation of miRNA targets. Methods. 2008;44:47–54. doi: 10.1016/j.ymeth.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5' UTR as in the 3' UTR. Proc. Natl Acad. Sci. USA. 2007;104:9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kong YW, Cannell IG, de Moor CH, Hill K, Garside PG, Hamilton TL, Meijer HA, Dobbyn HC, Stoneley M, Spriggs KA, et al. The mechanism of micro-RNA-mediated translation repression is determined by the promoter of the target gene. Proc. Natl Acad. Sci. USA. 2008;105:8866–8871. doi: 10.1073/pnas.0800650105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hendrickson DG, Hogan DJ, McCullough HL, Myers JW, Herschlag D, Ferrell JE, Brown PO. Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS Biol. 2009;7:e1000238. doi: 10.1371/journal.pbio.1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 46.Llave C, Xie Z, Kasschau KD, Carrington JC. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297:2053–2056. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- 47.Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjarn M, Hansen HF, Berger U, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 48.Elmen J, Lindow M, Silahtaroglu A, Bak M, Christensen M, Lind-Thomsen A, Hedtjarn M, Hansen JB, Hansen HF, Straarup EM, et al. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 2008;36:1153–1162. doi: 10.1093/nar/gkm1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 50.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat. Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haraguchi T, Ozaki Y, Iba H. Vectors expressing efficient RNA decoys achieve the long-term suppression of specific microRNA activity in mammalian cells. Nucleic Acids Res. 2009;37:e43. doi: 10.1093/nar/gkp040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bracken CP, Gregory PA, Kolesnikoff N, Bert AG, Wang J, Shannon MF, Goodall GJ. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008;68:7846–7854. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]
- 53.Hobert O. miRNAs play a tune. Cell. 2007;131:22–24. doi: 10.1016/j.cell.2007.09.031. [DOI] [PubMed] [Google Scholar]
- 54.Khan AA, Betel D, Miller ML, Sander C, Leslie CS, Marks DS. Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nat. Biotechnol. 2009;27:549–555. doi: 10.1038/nbt.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vinther J, Hedegaard MM, Gardner PP, Andersen JS, Arctander P. Identification of miRNA targets with stable isotope labeling by amino acids in cell culture. Nucleic Acids Res. 2006;34:e107. doi: 10.1093/nar/gkl590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tuddenham L, Wheeler G, Ntounia-Fousara S, Waters J, Hajihosseini MK, Clark I, Dalmay T. The cartilage specific microRNA-140 targets histone deacetylase 4 in mouse cells. FEBS Lett. 2006;580:4214–4217. doi: 10.1016/j.febslet.2006.06.080. [DOI] [PubMed] [Google Scholar]
- 57.Karginov FV, Conaco C, Xuan Z, Schmidt BH, Parker JS, Mandel G, Hannon GJ. A biochemical approach to identifying microRNA targets. Proc. Natl Acad. Sci. USA. 2007;104:19291–19296. doi: 10.1073/pnas.0709971104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu G, Fewell C, Taylor C, Deng N, Hedges D, Wang X, Zhang K, Lacey M, Zhang H, Yin Q, et al. Transcriptome and targetome analysis in MIR155 expressing cells using RNA-seq. RNA. 2010;16:1610–1622. doi: 10.1261/rna.2194910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Le Brigand K, Robbe-Sermesant K, Mari B, Barbry P. MiRonTop: mining microRNAs targets across large scale gene expression studies. Bioinformatics. 2010;26:3131–3132. doi: 10.1093/bioinformatics/btq589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Beitzinger M, Peters L, Zhu JY, Kremmer E, Meister G. Identification of human microRNA targets from isolated argonaute protein complexes. RNA Biol. 2007;4:76–84. doi: 10.4161/rna.4.2.4640. [DOI] [PubMed] [Google Scholar]
- 63.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr, Jungkamp AC, Munschauer M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Matkovich SJ, Van Booven DJ, Eschenbacher WH, Dorn GW., 2nd RISC RNA Sequencing for Context-Specific Identification of In Vivo MicroRNA Targets. Circ. Res. 2010;108:18–26. doi: 10.1161/CIRCRESAHA.110.233528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mili S, Steitz JA. Evidence for reassociation of RNA-binding proteins after cell lysis: implications for the interpretation of immunoprecipitation analyses. RNA. 2004;10:1692–1694. doi: 10.1261/rna.7151404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zisoulis DG, Lovci MT, Wilbert ML, Hutt KR, Liang TY, Pasquinelli AE, Yeo GW. Comprehensive discovery of endogenous Argonaute binding sites in Caenorhabditis elegans. Nat. Struct. Mol. Biol. 2010;17:173–179. doi: 10.1038/nsmb.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Orom UA, Lund AH. Isolation of microRNA targets using biotinylated synthetic microRNAs. Methods. 2007;43:162–165. doi: 10.1016/j.ymeth.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 68.Orom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5'UTR of ribosomal protein mRNAs and enhances their translation. Mol. Cell. 2008;30:460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 69.Gregory BD, O’Malley RC, Lister R, Urich MA, Tonti-Filippini J, Chen H, Millar AH, Ecker JR. A link between RNA metabolism and silencing affecting Arabidopsis development. Dev. Cell. 2008;14:854–866. doi: 10.1016/j.devcel.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 70.Addo-Quaye C, Miller W, Axtell MJ. CleaveLand: a pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics. 2009;25:130–131. doi: 10.1093/bioinformatics/btn604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.German MA, Luo S, Schroth G, Meyers BC, Green PJ. Construction of Parallel Analysis of RNA Ends (PARE) libraries for the study of cleaved miRNA targets and the RNA degradome. Nat. Protoc. 2009;4:356–362. doi: 10.1038/nprot.2009.8. [DOI] [PubMed] [Google Scholar]
- 72.Li YF, Zheng Y, Addo-Quaye C, Zhang L, Saini A, Jagadeeswaran G, Axtell MJ, Zhang W, Sunkar R. Transcriptome-wide identification of microRNA targets in rice. Plant J. 2010;62:742–759. doi: 10.1111/j.1365-313X.2010.04187.x. [DOI] [PubMed] [Google Scholar]
- 73.Eckardt NA. Investigating translational repression by microRNAs in Arabidopsis. Plant Cell. 2009;21:1624. doi: 10.1105/tpc.109.210613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vatolin S, Navaratne K, Weil RJ. A novel method to detect functional microRNA targets. J. Mol. Biol. 2006;358:983–996. doi: 10.1016/j.jmb.2006.02.063. [DOI] [PubMed] [Google Scholar]
- 75.Andachi Y. A novel biochemical method to identify target genes of individual microRNAs: identification of a new Caenorhabditis elegans let-7 target. RNA. 2008;14:2440–2451. doi: 10.1261/rna.1139508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang Y, Chaerkady R, Kandasamy K, Huang TC, Selvan LD, Dwivedi SB, Kent OA, Mendell JT, Pandey A. Identifying targets of miR-143 using a SILAC-based proteomic approach. Mol. Biosyst. 2010;6:1873–1882. doi: 10.1039/c004401f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhu S, Si ML, Wu H, Mo YY. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1) J. Biol. Chem. 2007;282:14328–14336. doi: 10.1074/jbc.M611393200. [DOI] [PubMed] [Google Scholar]
- 78.Muniyappa MK, Dowling P, Henry M, Meleady P, Doolan P, Gammell P, Clynes M, Barron N. MiRNA-29a regulates the expression of numerous proteins and reduces the invasiveness and proliferation of human carcinoma cell lines. Eur. J. Cancer. 2009;45:3104–3118. doi: 10.1016/j.ejca.2009.09.014. [DOI] [PubMed] [Google Scholar]
- 79.Wu S, Huang S, Ding J, Zhao Y, Liang L, Liu T, Zhan R, He X. Multiple microRNAs modulate p21Cip1/Waf1 expression by directly targeting its 3' untranslated region. Oncogene. 2010;29:2302–2308. doi: 10.1038/onc.2010.34. [DOI] [PubMed] [Google Scholar]
- 80.Vo NK, Dalton RP, Liu N, Olson EN, Goodman RH. Affinity purification of microRNA-133a with the cardiac transcription factor, Hand2. Proc. Natl Acad. Sci. USA. 2010;107:19231–19236. doi: 10.1073/pnas.1013162107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jiang Q, Feng MG, Mo YY. Systematic validation of predicted microRNAs for cyclin D1. BMC Cancer. 2009;9:194. doi: 10.1186/1471-2407-9-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Peter ME. Targeting of mRNAs by multiple miRNAs: the next step. Oncogene. 2010;29:2161–2164. doi: 10.1038/onc.2010.59. [DOI] [PubMed] [Google Scholar]