Abstract
1,3-Butadiene (BD) is a common environmental contaminant classified as “carcinogenic to humans.” Formation of BD-induced DNA adducts plays a major role in its carcinogenicity. BD is also an epigenotoxic agent (i.e., it affects DNA and histone methylation in the liver). We used a panel of genetically diverse inbred mice (NOD/LtJ, CAST/EiJ, A/J, WSB/EiJ, PWK/PhJ, C57BL/6J, and 129S1/SvImJ) to assess whether BD-induced genotoxic and epigenotoxic events may be subject to interstrain differences. Mice (male, 7 weeks) were exposed via inhalation to 0 or 625 ppm BD for 6 h/day and 5 days/week for 2 weeks and liver BD-DNA adducts, epigenetic alterations, and liver toxicity were assessed. N-7-(2,3,4-trihydroxybut-1-yl)-guanine adducts were detected in all strains after exposure, yet BD-induced DNA damage in CAST/EiJ mice was two to three times lower. Epigenetic effects of BD were most prominent in C57BL/6J mice where loss of global DNA methylation and loss of trimethylation of histone H3 lysine 9, histone H3 lysine 27, and histone H4 lysine 20, accompanied by dysregulation of liver gene expression indicative of hepatotoxicity, were found. Interestingly, we observed an increase in histone methylation in the absence of changes in gene expression and DNA methylation in CAST/EiJ strain. We hypothesized that mitigated genotoxicity of BD in CAST/EiJ mice may be due to chromatin condensation. Indeed, we show that in response to BD exposure, chromatin condensation occurs in CAST/EiJ, whereas the opposite effect is observed in C57BL/6J mice. These findings demonstrate that interstrain susceptibility to genotoxicity by a well-known environmental carcinogen may be due to strain-specific epigenetic events in response to the exposure.
Keywords: liver, systems toxicology, 1,3-butadiene; mouse, epigenetics
Environmental and occupational exposures to man-made chemical agents, in spite of the major efforts for increased regulatory action in both United States and Europe, continue to be a major public health concern worldwide (Judson et al., 2009; Lucier and Schecter, 1998; Ziegler, 1993). It is widely believed that exposure to chemicals is one of the major causes of many human diseases (Rappaport and Smith, 2010), including cancer (Loeb and Harris, 2008; Wild, 2009). There are a number of major research efforts that are directed at advancing the science of rapid identification, assessment, and mitigation of exposures to hazardous chemical agents, especially agents with a carcinogenic potential (Kavlock et al., 2009). Recent advances in the field of research on the mechanisms of toxicity and carcinogenicity indicate that a wide range of chemical carcinogens, in addition to inducing genetic changes, affect and alter the cellular epigenetic state (Pogribny et al., 2008; Wild, 2009). These discoveries redefine the classical model of multistage carcinogenesis to include both genetic and epigenetic changes at each stage of the carcinogenic process (Jones and Baylin, 2007).
1,3-Butadiene (BD) is a major industrial chemical and a common environmental contaminant that is classified as carcinogenic to humans (IARC, 2009; Swenberg et al., 2010). BD is metabolized via oxidation by the family of cytochrome P450 mono-oxygenases to form reactive epoxides: 1,2-epoxy-3-butene, 1,2:3,4-diepoxybutane, and 3,4-epoxy-1,2-butanediol (Filser et al., 2007; Himmelstein et al., 1996). These secondary metabolites may directly interact with DNA and form mutagenic DNA adducts (Cochrane and Skopek, 1994; Kemper et al., 2001). The covalent interaction of metabolic derivatives of BD with DNA is believed to be a critical step in the initiation of tumorigenesis (Poirier, 2004; Swenberg et al., 2000). It was also shown recently that short-term exposure to BD also leads to a variety of nongenotoxic epigenetic alterations in the liver of C57BL/6J mice (Koturbash et al., 2011). These findings support the hypothesis that epigenetic alterations may serve as early indicators of exposure to agents with a carcinogenic potential and may be used as biomarkers in the assessment of hazardous agents (LeBaron et al., 2010; Marlowe et al., 2009).
The elucidation of the mechanisms of toxicity is usually carried out in genetically homogeneous in vivo or in vitro models in order to fix as many variables as possible. This provides information in a single strain or cell line, yet the extrapolation of such data to the population effects is constrained by the inference from a single genome to model complex human phenotypes. To address these limitations, novel animal models are needed. The need to “account for differences among humans in cancer susceptibility other than from possible early-life susceptibility” (National Research Council, 2008) is recognized by both the scientific community and the regulatory agencies. A multistrain approach provides critical information for understanding the genetic background-dependent and -independent components of chemical's mode of action, estimation of interindividual differences in toxicodynamics and –kinetics, and identification of biomarkers of toxicity (Rusyn et al., 2010). In this study, we show that important strain differences in both genotoxic and epigenotoxic responses to BD exist, and the chromatin condensation response is an underlying mechanism for such differences.
MATERIALS AND METHODS
Animals and experimental design.
Male CAST/EiJ, NOD/LtJ, A/J, WSB/EiJ, PWK/PhJ, 129S1/SvImJ, and C57BL/6J mice (7 weeks of age) were housed in a sterilized animal facility at the University of North Carolina at Chapel Hill in a temperature-controlled room with a 12-h light/dark cycle. These mice were from a colony of mice maintained at the University of North Carolina at Chapel Hill and used for the derivation of the Collaborative Cross (Aylor et al., 2011). The founders of the colony were originally obtained from the Jackson Laboratory (Bar Harbor, ME). These strains were selected because they provide an excellent representation of the genetic diversity present in the laboratory mouse by sampling the three main subspecies of house mouse (Yang et al., 2011). They are also seven of the eight founders of the Collaborative Cross (Chesler et al., 2008), and their genomes have been fully sequenced at least at ×20 coverage (http://www.sanger.ac.uk/resources/mouse/genomes/). Animals were given food (NIH-31 diet, Purina Mills, Richmond, IN) and water ad libitum.
Following the 2-week acclimation period, the mice were randomly assigned to control (n = 5 per strain, exposed to filtered air) or experimental (n = 5 per strain, exposed to 625 ppm of BD) groups. Exposure concentration and protocol were selected based on the National Toxicology Program's carcinogenesis studies with butadiene in mice (National Toxicology Program, 1984, 1993). Exposure was performed for 6 h/day and 5 days/week (Monday through Friday) for 2 weeks. On each experimental day, the mice were placed in a cylindrical metal mesh holder for the duration of BD exposure and then transferred back to their regular cages. Concentrations of BD in exposure chambers were monitored at the beginning and end of each exposure period using gas chromatography and were determined to correspond to the target concentrations. Mice were euthanized by exsanguination following deep isoflurane anesthesia after the last exposure to BD. The livers were excised, snap-frozen in liquid nitrogen, and stored at −80°C for subsequent analyses. All experimental procedures involving animals were reviewed and approved by the Institutional Animal Care and Use Committee at the University of North Carolina at Chapel Hill.
Determination of N-7 guanine adduct formation.
Genomic DNA was isolated from mouse liver tissues by standard digestion of DNA with proteinase K, followed by phenol-chloroform extraction and ethanol precipitation. The analysis of N-7-(2,3,4-trihydroxybut-1-yl)-guanine (THB-Gua) was performed following neutral thermal hydrolysis by liquid chromatography/positive ion electrospray ionization/mass spectrometry/mass spectrometry (LC/ESI+/MS/MS) as described elsewhere (Koc et al., 1999).
Determination of global DNA methylation status by methylation-sensitive cytosine extension assay.
The extent of global DNA methylation was evaluated with a [3H]dCTP extension assay as described previously (Pogribny et al., 1999).
Methylation-sensitive quantitative PCR analysis of long interspersed elements 1 and major and minor satellites repetitive elements.
The methylation status of long interspersed elements 1 (LINE1) and major and minor satellites repetitive elements was determined by methylation-sensitive McrBC-quantitative PCR (qPCR) assay (Martens et al., 2005). Briefly, genomic DNA (1 μg) was digested overnight with the methylation-specific restriction enzyme McrBC (New England Biolabs, Ipswich, MA) and then analyzed by qPCR on an Applied Biosystems (Forrest City, CA) 7900 Real-Time PCR System. The threshold cycle (Ct) is defined as the fractional cycle number that passes the fixed threshold. The Ct values were converted into the absolute amount of input DNA using the absolute standard curve method. An increased amount of input DNA after digestion with McrBC is indicative of hypomethylation, whereas a decreased amount of input DNA is indicative of hypermethylation.
Western blot analysis of histone modifications.
The status of histone H3 lysine 4 (H3K4), histone H3 lysine 9 (H3K9), histone H3 lysine 27 (H3K27), and histone H4 lysine 20 (H4K20) trimethylation in the livers of control and BD-exposed mice was determined by Western blot analysis (Tryndyak et al., 2006). All the antibodies were purchased from Millipore Corporation (Billerica, MA). Chemiluminescence detection was performed with the HRP substrate for Western blotting (Millipore Corporation) and measured directly by a BioSpectrum AC Imaging System (Upland, CA). The signal intensity was analyzed by ImageQuant software (Molecular Dynamics, Sunnyvale, CA).
Quantitative reverse transcription-PCR array analysis of the hepatotoxicity-related gene expression.
Total RNA was extracted from mouse liver tissues using TRI Reagent (Ambion, Austin, TX) according to the manufacturer's instructions. Complementary DNA (cDNA) was synthesized from 5 μg of total RNA using RT2 First-Strand cDNA Synthesis kit (SABiosciences, Frederick, MD). The Mouse Hepatotoxicity RT2 Profiler PCR Arrays (SABiosciences) were used according to the manufacturer's protocol to determine the expression of 84 key genes implicated as potential biomarkers of liver toxicity. The relative level of messenger RNA (mRNA) for each gene was determined using the 2ΔΔCt method (Livak and Schmittgen, 2001). The results are presented as fold change for each mRNA in the livers of mice.
Quantitative reverse transcription-PCR.
The levels of gene transcripts for histone lysine methyltransferases Suv39h1 (Mm00468952_m1), Prdm2 (Mm01348917_m1), and enhancer of zeste homolog 2, Drosophila (Ezh2, Mm00468464_m1) were determined by quantitative reverse transcription-PCR (qRT-PCR) using TaqMan Gene Expression Assays (Applied Biosystems) according to the manufacturer's protocol. Expression of repetitive elements in the mouse genome, including LINE1 and major and minor satellites, was determined by qRT-PCR (Martens et al., 2005).
Methylation-sensitive analysis of chromatin structure.
The chromatin structure in the livers was determined by a modified methylation-based analysis of nucleosomal DNA accessibility (Miranda et al., 2010). This technique is based on the fact that CpG sites in DNA are protected from methylation when these sequences are wrapped around histones (Kladde and Simpson, 1994; Miranda et al., 2010). Briefly, nuclei from 40 μg of liver tissue of control and BD-exposed mice were isolated and purified as described previously (Miranda et al., 2010). After purification, the nuclei were immediately resuspended in M.HpaII buffer (New England Biolabs, Ipswich, MA) and incubated with 60 U of HpaII methylase for 15 min at 37°C. The reaction was terminated by the addition of 150 μl of stop solution (10mM TrisCl, 300mM NaCl, 1% SDS, and 5mM disodium EDTA), and genomic DNA was extracted using the standard digestion with proteinase K, followed by phenol/chlolorophorm/isoamyl alcohol extraction and ethanol precipitation (Strauss, 1989). The extent of CCGG methylation was evaluated with a [3H]dCTP extension assay (Pogribny et al., 1999).
Statistical analyses.
Results are presented as mean ± SD. Statistical analyses were conducted by two-way ANOVA, with pairwise comparisons being made by the Student-Newman-Keuls method. When necessary, the data were natural log-transformed before conducting the analyses to maintain a more equal variance or normal data distribution. p Values < 0.05 were considered significant.
RESULTS
Hepatic N-7 Guanine Adduct Levels
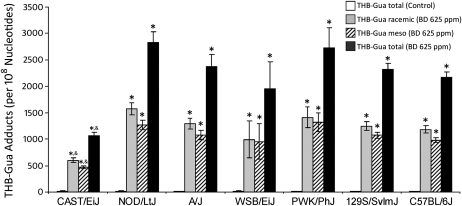
LC/ESI+/MS/MS was used to assess the amount of THB-Gua adducts in hepatic DNA of control mice and each strain of mice exposed to 625-ppm BD. Inhalational exposure to BD resulted in extensive formation of racemic and meso THB-Gua adducts in hepatic DNA in all strains tested (Fig. 1). In addition, the THB-Gua adducts in liver DNA of BD-exposed NOD/LtJ, A/J, WSB/EiJ, PWK/PhJ, 129S1/SvImJ, and C57BL/6J mice were two- to threefold higher than the adducts found in the livers of BD-exposed CAST/EiJ mice, differences that were significant.
FIG. 1.
Amounts of THB-Gua-BD adducts in liver DNA from mice exposed to 0- or 625-ppm BD. Data are presented as mean ± SD (n = 5). Asterisk and ampersand (* and &) denote significant (p < 0.05) differences in the same DNA adduct as compared with the corresponding strain's control mice or as compared with BD-treated mice of other strains, respectively.
Effect of BD Exposure on Global DNA Methylation and Methylation of the Repetitive DNA Elements
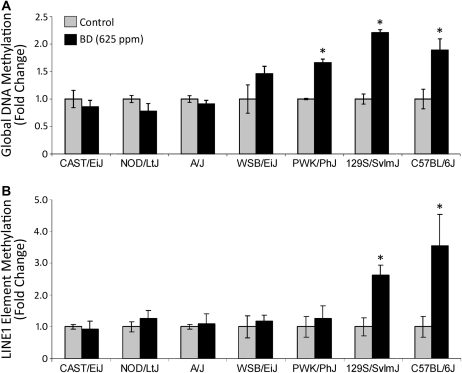
The extent of interstrain differences in epigenetic responses to BD treatment was also evaluated. First, we evaluated DNA methylation, both global and LINE1 repetitive elements, in the livers of BD-exposed and control mice. Figure 2 shows that exposure to BD caused strain-dependent changes in the extent of hepatic DNA methylation. Specifically, levels of global and LINE1 DNA methylation were significantly decreased after BD exposure in 129S1/SvImJ and C57BL/6J mice. In PWK/PhJ mice, BD led to a significant decrease in global DNA methylation only. No effects were observed in other (CAST/EiJ, NOD/LtJ, A/J, and WSB/EiJ) strains.
FIG. 2.
Effects of BD exposure on the extent of DNA methylation in mouse liver. (A) Loss of global DNA methylation in the livers of BD-exposed mice as measured by a cytosine extension ([3H]dCTP incorporation) DNA methylation assay. (B) Loss of LINE1 repetitive elements methylation in the livers of BD-exposed mice as measured by a methylation-sensitive McrBC-qPCR assay. Data are presented as fold change in BD (625 ppm)-exposed mice relative to the control mice in each strain, mean ± SD (n = 5). Asterisks (*) denote a significant (p < 0.05) difference from the corresponding strain's control group.
Effect of BD Exposure on Histone Lysine Methylation
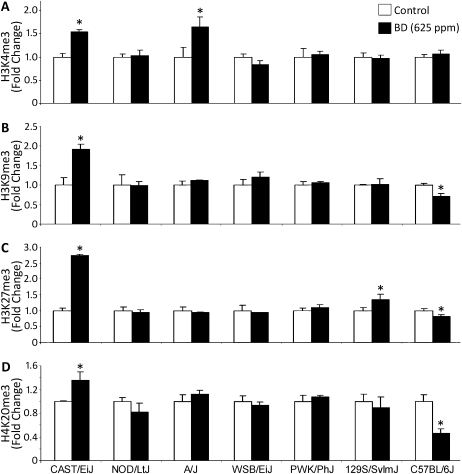
Previously we observed that BD exposure also affects lysine methylation in liver histones in C57BL/6J mice, specifically decreases in trimethylation of histone H3K9, H3K27, and H4K20 (Koturbash et al., 2011). Although in C57BL/6J strain the changes in DNA and histone methylation were both prominent and concordant, lack of BD effect on DNA methylation in other strains led us to examine the status of these key repressive histone marks in the livers of other BD-exposed strains. Figure 3 shows that exposure to BD had little effect on histone methylation in other strains examined, with the exception of a significant increase in H3K4me3, H3K9me3, H3K27me3, and H4K20me3 in CAST/EiJ mice. Specifically, the levels of H3K4, H3K9, H3K27, and H4K20 trimethylation in the livers of BD-exposed CAST/EiJ mice were 1.5, 1.9, 2.7, and 1.4 times higher, respectively, as compared with the age-matched control CAST/EiJ mice. Level of H3K4me3 was also elevated in BD-exposed A/J mice. A small, but significant, increase in H3K27me3 was also observed in BD-treated 129S1/SvlmJ mice.
FIG. 3.
Effects of BD exposure on histone lysine trimethylation in mouse liver. Histone H3K4me3 (A), histone H3K9me3 (B), histone H3K27me3 (C), and histone H4K20me3 (D) levels were assessed by immunostaining using specific antibodies against trimethylated histones. Equal sample loading was confirmed by immunostaining against total histone H3 or H4 where appropriate (data not shown). Densitometry analysis of the immunostaining results is shown as change in methylation levels relative to control after correction for the total amount of each histone in the individual samples. Data are presented as mean ± SD (n = 5). Asterisks (*) denote a significant (p < 0.05) difference from the corresponding strain's control group.
Effect of BD Exposure on Expression of Hepatotoxicity-Related Biomarker Genes
Based on the observation of major strain-specific differences in both genotoxic and epigenotoxic effects of BD, we selected C57BL/6J, CAST/EiJ, and A/J strains, as representative of hypo-, hyper-, or no effect of BD on the epigenome, for further analyses. It is widely believed that a covalent interaction of metabolic reactive intermediates of genotoxic carcinogens, including those formed from BD, with DNA is a critical event in the initiation of carcinogenesis (Swenberg et al., 2000); however, although necessary, these genotoxic effects are not sufficient to explain tumor development. It is believed that hepatotoxicity also plays a role in liver carcinogenesis (Neumann, 2009); therefore we determined the expression of genes considered to be markers of hepatotoxicity (Gao et al., 2010) in three strains of mice (CAST/EiJ, A/J, and C57BL/6J) that differed in their genotoxic and epigenetic responses to BD exposure.
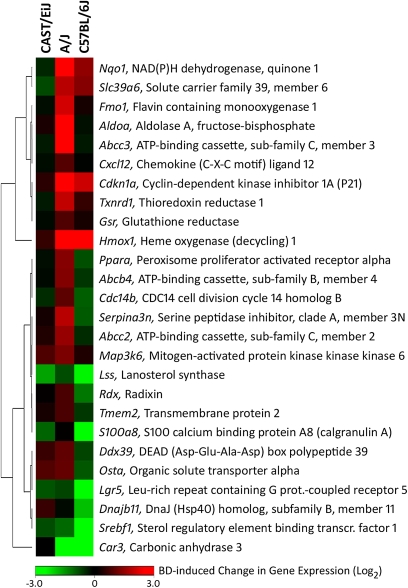
Although no change in serum alanine aminotransferase activity was observed in mice exposed to 625-ppm BD (data not shown), histopathological evaluation of the liver sections revealed that the hepatocytes of the BD-treated C57BL/6J mice, but not of any other strain examined, showed diffused glycogen depletion characterized by the lack of cytoplasmic vacuoles (Koturbash et al., 2011). Gene expression analysis revealed that exposure of C57BL/6J mice to BD caused marked changes in expression of the majority of genes, including Hmox1, Nqo1, Car3, Srebf1, and Lgr5, response indicative of liver injury (Fig. 4, Supplementary table 1). Changes in expression of these genes were also observed in the livers of BD-exposed A/J mice, albeit they were less pronounced. Upregulation of genes that belong to the ATP-binding cassette (ABC) transporters, including Abcc2, Abcc4, Abbc3, and Abcb11, was found in the livers of A/J mice exposed to BD. Products of these genes are transmembrane proteins that actively export various substrates and metabolic intermediates and products from the liver cells to the bile. Interestingly, few gene expression changes indicative of hepatotoxicity were detected in the liver tissue of BD-exposed CAST/EiJ mice.
FIG. 4.
Heatmap of differentially expressed hepatotoxicity biomarker genes in the livers of control and BD-exposed C57BL/6J, A/J, and CAST/EiJ mice. Gene expression was determined in total liver RNA from control and BD-exposed mice (n = 3 from each group). A heatmap (average of fold change between control and exposed groups within each strain) of genes identified as significant (p < 0.05) and changing in expression by at least 1.5-fold is shown. The color bar identifies up and downregulated genes as indicated. See Supplementary table 1 for a list of genes and gene expression values.
Analysis of Chromatin Structure
Next, we hypothesized that the observed strain-specific differences in the effects of BD on liver histone and DNA methylation, transcriptional response, and DNA damage may be indicative of dissimilarities of the effects on chromatin structure. This hypothesis was based on the observation that the level of background histone methylation in the liver varies significantly between CAST/EiJ, A/J, and C57BL/6J strains (Supplementary fig. 1).
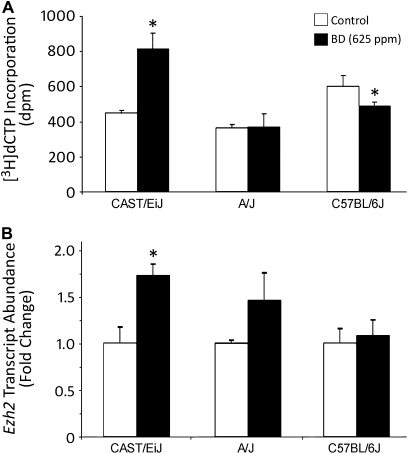
It is well-established that CpG sites within DNA are protected from methylation when these sequences are wrapped around histones (Kladde and Simpson, 1994; Miranda et al., 2010). Therefore, CpG sites in condensed heterochromatin are inaccessible for methylation, whereas CpG sites in relaxed chromatin are accessible for methylation. It has been reported that nearly 90% of HpaII sequences (CCGG) in the mouse genome are located outside of CpG islands, predominantly within transposable elements (Fazzari and Greally, 2004). Thus, we analyzed the structure of hepatic chromatin in control and BD-exposed CAST/EiJ, A/J, and C57BL/6J mice by determining the accessibility of nucleosomal DNA to methylation of HpaII sites. Figure 5A shows that exposure of CAST/EiJ mice to BD resulted in formation of a more condensed chromatin. This was evidenced by a nearly twofold greater incorporation of [3H]dCTP into HpaII-digested DNA from HpaII methylase-pretreated nuclei isolated from the livers of BD-exposed CAST/EiJ mice as compared with control mice. In contrast, a decrease in [3H]dCTP incorporation into DNA after pretreatment of nuclei from the livers of BD-treated C57BL/6J mice with HpaII methylase indicated the formation of more open, relaxed chromatin and, therefore, increased accessibility of CCGG sites to methylation. Interestingly, BD exposure did not have an effect on chromatin structure in A/J mice.
FIG. 5.
Analysis of chromatin structure in the livers of control and BD-exposed C57BL/6J, A/J, and CAST/EiJ mice. (A) Chromatin structure in the livers of control and BD-exposed C57BL/6J, A/J, and CAST/EiJ mice was determined by analyzing the accessibility of CCGG sites within nucleosomal DNA to methylation. The extent of [3H]dCTP incorporation into DNA is directly proportional to changes in chromatin condensation. (B) Total liver RNA from control mice (white bars) and mice exposed to 625-ppm BD (black bars) was used to evaluate transcript abundance of Ezh2. Data are presented as mean ± SD (n = 5). Asterisks (*) denote a significant (p < 0.05) difference from the corresponding strain's control group.
Ezh2 is a key histone lysine methyltransferase that catalyzes the methylation of nucleosomal histone H3 at lysine 27 (Cao et al., 2002). We tested whether BD exposure has an effect on the expression of Ezh2. Figure 5B shows that exposure to BD resulted in significant upregulation of Ezh2 in the livers of CAST/EiJ mice only. No BD effect on histone methyltransferases, Suv39h1 and Prdm2, was found.
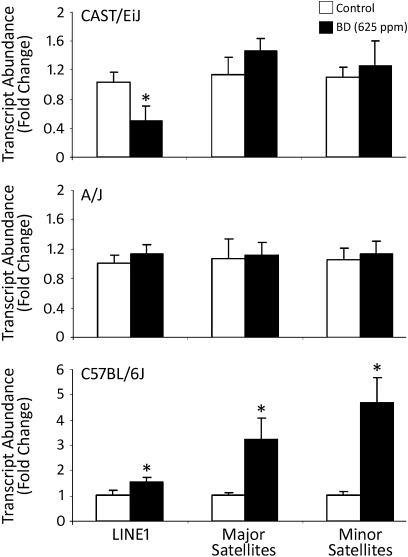
The formation of different chromatin structures in the livers of CAST/EiJ and C57BL/6J mice exposed to BD was further confirmed by the analysis of expression of LINE1 and major and minor satellites, main repetitive elements in the mouse genome. Figure 6 shows an approximate twofold decrease in the level of LINE1 expression in BD-exposed CAST/EiJ mice (top panel), whereas expression of LINE1 and major and minor satellites in the livers of BD-exposed C57BL/6J mice was 1.5, 3.2, and 4.7 times greater, respectively (bottom panel), as compared with control mice. No changes in LINE1 and major and minor satellites expression were found in the livers of BD-exposed A/J mice (middle panel).
FIG. 6.
Expression of LINE1 and major and minor satellites in the livers of control and BD-exposed CAST/EiJ, A/J, and C57BL/6J mice. Total liver RNA from control mice (white bars) and mice exposed to 625-ppm BD (black bars) was used to evaluate transcript abundance of LINE1, as well as major and minor satellites. Data are presented as mean ± SD (n = 5). Asterisks (*) denote a significant (p < 0.05) difference from the corresponding strain's control group.
DISCUSSION
The results of the present study demonstrate that short-term inhalational exposure of mice to the environmental contaminant and potent carcinogen, BD, is characterized by substantial differences in hepatic genetic and epigenetic response among mouse strains. Specifically, although formation of extensive amounts of THB-Gua adducts was observed in the livers of mice from all seven BD-exposed inbred strains, DNA damage was significantly less in CAST/EiJ strain (Fig. 1). Our previous study demonstrated that the presence of THB-Gua adducts in the livers of BD-exposed C57BL/6J mice is associated with disruption in the hepatic epigenetic status (Koturbash et al., 2011). The present study shows that BD exposure, similarly to the observed differences in genotoxicity response, resulted in substantial variability in epigenetic alterations among strains. Even though many reports have demonstrated strain-specific variability in toxicity in response to chemical exposure (Bradford et al., 2011; Harrill et al., 2009a,b), a full understanding of the underlying mechanisms of such differences remains unresolved.
A detailed analysis of the pattern of genetic and epigenetic alterations caused by BD exposure allowed us to group mouse strains into three categories: (1) mice with high levels of THB-Gua adducts (Fig. 1) and a substantial decrease in global and/or LINE1 DNA methylation (WSB/EiJ, PWK/PhJ, 129S1/SvImJ, and C57BL/6J strains; Fig. 2); C57BL/6J mice were also characterized by a substantial decrease in histone H3K9, H3K27, and H4K20 trimethylation (Fig. 3); (2) mice with high levels of THB-Gua adducts and no significant alterations in global or LINE1 DNA methylation (NOD/LtJ and A/J strains); and (3) mice with low levels of hepatic THB-Gua adducts and increased histone lysine methylation (CAST/EiJ strain). By selecting representative strains from each group, we showed that the strain-specific differences may be due to dissimilarities in chromatin structure in response to a genotoxic insult.
It is well-established that both DNA methylation and histone H3K9, H3K27, and H4K20 trimethylation play a crucial role in the maintenance of proper chromatin structure and genomic stability (Dillon, 2004; Jenuwein, 2006; Martin and Zhang, 2005). For instance, a greater extent of DNA methylation and histone H3K9, H3K27, and H4K20 trimethylation are associated with condensed chromatin structure and, consequently, gene silencing. In contrast, decreased DNA and histone H3K9, H3K27, and H4K20 methylation is associated with the formation of relaxed chromatin, increased transcription of the LINE1 and other repetitive DNA sequences, and a variety of genomic instability events. Additionally, it has been suggested that chromatin structure may influence the sensitivity of DNA to damage caused by various environmental agents (Falk et al., 2008).
The results of our study support the hypothesis that interstrain differences in response to BD exposure may be caused by alterations in chromatin structure. A profound loss of DNA and histone H3K9, H3K27, and H4K20 methylation, a relaxation of chromatin structure, activation of expression of LINE1 and major and minor satellites, and liver toxicity in BD-exposed C57BL/6J mice support this suggestion. In contrast, in the livers of A/J mice exposed to BD, the hepatotoxic effect of BD was substantially lower, despite comparable amounts of THB-Gua adducts. This may be explained, in part, by a substantial upregulation of ABC-transporter genes and the ability of liver cells to retain a proper epigenetic status.
The most intriguing findings of our study are the substantially lower formation of THB-Gua adducts and negligible hepatotoxicity of BD exposure in CAST/EiJ mice. As mentioned above, CAST/EiJ was the only strain that upon BD exposure exhibited a substantial increase in histone H3K9, H3K4, H4K20, and especially H3K27 trimethylation. These observations were supported by an increase in expression of the Ezh2 histone methyltransferase. These changes resulted in formation of a compact heterochromatin structure that may decrease accessibility of DNA to BD-reactive intermediates yielding lower formation of THB-Gua adducts and less liver toxicity. In addition, increased capacity for detoxication/elimination or lower capacity of bioactivation of BD in CAST/EiJ strain, which may well be due to epigenetic modulation of gene expression, may also play a role.
The linkages between environmental agent-induced DNA damage, epigenetic effects, and subsequent risk of mutagenesis remain largely unexplored; however, there is increasing evidence that chromatin structure strongly influences DNA repair processes (Kinner et al., 2008). Our data show that BD-induced DNA and histone hypomethylation effects that have been associated with carcinogenesis through spontaneous mutations, including loss of heterozygosity, chromosome translocation, and DNA deletion (Chen et al., 1998), may lead to increased cancer susceptibility in some, but not all strains. In addition, strain-specific changes in chromatin structure may not only prevent chemical-induced DNA damage but also impact DNA repair because replication-independent endogenous DNA double-strand breaks are processed differently in methylated and unmethylated parts of chromatin (Kongruttanachok et al., 2010). The relevance of the repair pathways to these strain-dependent differences remains to be elucidated because THB-Gua adducts are primarily lost through chemical depurination.
In conclusion, our findings demonstrate that significant epigenetic events occur following BD exposure and suggest that the differences in interindividual susceptibility to BD and other genotoxic chemicals may be determined by variations in epigenetic response. More importantly, the results of the present study suggest that assessment of carcinogen-induced epigenetic alterations, in addition to genetic changes, may facilitate identification of subpopulation susceptible to exposure.
FUNDING
National Institutes of Health (R01 ES012689, R01 ES015241, and P30 ES010126).
Acknowledgments
The views expressed in this paper do not necessarily represent those of the U.S. Food and Drug Administration.
References
- Aylor DL, Valdar W, Foulds-Mathes W, Buus RJ, Verdugo RA, Baric RS, Ferris MT, Frelinger JA, Heise M, Frieman MB, et al. Genetic analysis of complex traits in the emerging collaborative cross. Genome Res. Advance Access published on March 15, 2011; doi:10.1101/gr.111310.110. 2011 doi: 10.1101/gr.111310.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford BU, Lock EF, Kosyk O, Kim S, Uehara T, Harbourt D, Desimone M, Threadgill DW, Tryndyak V, Pogribny IP, et al. Inter-strain differences in the liver effects of trichloroethylene in a multi-strain panel of inbred mice. Toxicol. Sci. 2011;120:206–217. doi: 10.1093/toxsci/kfq362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Chen RZ, Pettersson U, Beard C, Jackson-Grusby L, Jaenisch R. DNA hypomethylation leads to elevated mutation rates. Nature. 1998;395:89–93. doi: 10.1038/25779. [DOI] [PubMed] [Google Scholar]
- Chesler EJ, Miller DR, Branstetter LR, Galloway LD, Jackson BL, Philip VM, Voy BH, Culiat CT, Threadgill DW, Williams RW, et al. The collaborative cross at Oak Ridge National Laboratory: developing a powerful resource for systems genetics. Mamm. Genome. 2008;19:382–389. doi: 10.1007/s00335-008-9135-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochrane JE, Skopek TR. Mutagenicity of butadiene and its epoxide metabolites: I. Mutagenic potential of 1,2-epoxybutene, 1,2,3,4-diepoxybutane and 3,4-epoxy-1,2-butanediol in cultured human lymphoblasts. Carcinogenesis. 1994;15:713–717. doi: 10.1093/carcin/15.4.713. [DOI] [PubMed] [Google Scholar]
- Dillon N. Heterochromatin structure and function. Biol. Cell. 2004;96:631–637. doi: 10.1016/j.biolcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Falk M, Lukasova E, Kozubek S. Chromatin structure influences the sensitivity of DNA to gamma-radiation. Biochim. Biophys. Acta. 2008;1783:2398–2414. doi: 10.1016/j.bbamcr.2008.07.010. [DOI] [PubMed] [Google Scholar]
- Fazzari MJ, Greally JM. Epigenomics: beyond CpG islands. Nat. Rev. Genet. 2004;5:446–455. doi: 10.1038/nrg1349. [DOI] [PubMed] [Google Scholar]
- Filser JG, Hutzler C, Meischner V, Veereshwarayya V, Csanady GA. Metabolism of 1,3-butadiene to toxicologically relevant metabolites in single-exposed mice and rats. Chem. Biol. Interact. 2007;166:93–103. doi: 10.1016/j.cbi.2006.03.002. [DOI] [PubMed] [Google Scholar]
- Gao W, Mizukawa Y, Nakatsu N, Minowa Y, Yamada H, Ohno Y, Urushidani T. Mechanism-based biomarker gene sets for glutathione depletion-related hepatotoxicity in rats. Toxicol. Appl. Pharmacol. 2010;247:211–221. doi: 10.1016/j.taap.2010.06.015. [DOI] [PubMed] [Google Scholar]
- Harrill AH, Ross PK, Gatti DM, Threadgill DW, Rusyn I. Population-based discovery of toxicogenomics biomarkers for hepatotoxicity using a laboratory strain diversity panel. Toxicol. Sci. 2009a;110:235–243. doi: 10.1093/toxsci/kfp096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrill AH, Watkins PB, Su S, Ross PK, Harbourt DE, Stylianou IM, Boorman GA, Russo MW, Sackler RS, Harris SC, et al. Mouse population-guided resequencing reveals that variants in CD44 contribute to acetaminophen-induced liver injury in humans. Genome Res. 2009b;19:1507–1515. doi: 10.1101/gr.090241.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelstein MW, Turner MJ, Asgharian B, Bond JA. Metabolism of 1,3-butadiene: inhalation pharmacokinetics and tissue dosimetry of butadiene epoxides in rats and mice. Toxicology. 1996;113:306–309. doi: 10.1016/0300-483x(96)03462-2. [DOI] [PubMed] [Google Scholar]
- IARC. 1,3-Butadiene, Ethylene Oxide and Vinyl Halides (Vinyl Fluoride and Vinyl Bromide) Lyon, France: WHO; 2009. [Google Scholar]
- Jenuwein T. The epigenetic magic of histone lysine methylation. FEBS J. 2006;273:3121–3135. doi: 10.1111/j.1742-4658.2006.05343.x. [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson R, Richard A, Dix DJ, Houck K, Martin M, Kavlock R, Dellarco V, Henry T, Holderman T, Sayre P, et al. The toxicity data landscape for environmental chemicals. Environ. Health Perspect. 2009;117:685–695. doi: 10.1289/ehp.0800168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavlock RJ, Austin CP, Tice RR. Toxicity testing in the 21st century: implications for human health risk assessment. Risk Anal. 2009;29:485–487. doi: 10.1111/j.1539-6924.2008.01168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemper RA, Krause RJ, Elfarra AA. Metabolism of butadiene monoxide by freshly isolated hepatocytes from mice and rats: different partitioning between oxidative, hydrolytic, and conjugation pathways. Drug Metab. Dispos. 2001;29:830–836. [PubMed] [Google Scholar]
- Kinner A, Wu W, Staudt C, Iliakis G. Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res. 2008;36:5678–5694. doi: 10.1093/nar/gkn550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kladde MP, Simpson RT. Positioned nucleosomes inhibit Dam methylation in vivo. Proc. Natl. Acad. Sci. U.S.A. 1994;91:1361–1365. doi: 10.1073/pnas.91.4.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koc H, Tretyakova NY, Walker VE, Henderson RF, Swenberg JA. Molecular dosimetry of N-7 guanine adduct formation in mice and rats exposed to 1,3-butadiene. Chem. Res. Toxicol. 1999;12:566–574. doi: 10.1021/tx980265f. [DOI] [PubMed] [Google Scholar]
- Kongruttanachok N, Phuangphairoj C, Thongnak A, Ponyeam W, Rattanatanyong P, Pornthanakasem W, Mutirangura A. Replication independent DNA double-strand break retention may prevent genomic instability. Mol. Cancer. 2010;9:70. doi: 10.1186/1476-4598-9-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koturbash I, Scherhag A, Sorrentino J, Sexton K, Bodnar W, Tryndyak V, Latendresse JR, Swenberg JA, Beland FA, Pogribny IP, et al. Epigenetic alterations in liver of C57BL/6J mice after short-term inhalational exposure to 1,3-butadiene. Environ. Health Perspect. 2011;119:635–640. doi: 10.1289/ehp.1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBaron MJ, Rasoulpour RJ, Klapacz J, Ellis-Hutchings RG, Hollnagel HM, Gollapudi BB. Epigenetics and chemical safety assessment. Mutat. Res. 2010;705:83–95. doi: 10.1016/j.mrrev.2010.04.003. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Loeb LA, Harris CC. Advances in chemical carcinogenesis: a historical review and prospective. Cancer Res. 2008;68:6863–6872. doi: 10.1158/0008-5472.CAN-08-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucier GW, Schecter A. Human exposure assessment and the National Toxicology Program. Environ. Health Perspect. 1998;106:623–627. doi: 10.1289/ehp.106-1533173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlowe J, Teo SS, Chibout SD, Pognan F, Moggs J. Mapping the epigenome–impact for toxicology. EXS. 2009;99:259–288. doi: 10.1007/978-3-7643-8336-7_10. [DOI] [PubMed] [Google Scholar]
- Martens JH, O'Sullivan RJ, Braunschweig U, Opravil S, Radolf M, Steinlein P, Jenuwein T. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J. 2005;24:800–812. doi: 10.1038/sj.emboj.7600545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- Miranda TB, Kelly TK. Bouazoune K, Jones PA. Methylation-sensitive single-molecule analysis of chromatin structure. Curr. Protoc. Mol. Biol. doi: 10.1002/0471142727.mb2117s89. Chapter 21, Unit 21.17.1--21.17.16. [DOI] [PubMed] [Google Scholar]
- National Research Council. Science and Decisions: Advancing Risk Assessment. Washington, DC: The National Academies Press; 2008. [PubMed] [Google Scholar]
- National Toxicology Program. NTP toxicology and carcinogenesis studies of 1,3-butadiene (CAS No. 106-99-0) in B6C3F1 mice (inhalation studies) Natl. Toxicol. Program Tech. Rep. Ser. 1984;288:1–111. [PubMed] [Google Scholar]
- National Toxicology Program. NTP toxicology and carcinogenesis studies of 1,3-butadiene (CAS No. 106-99-0) in B6C3F1 mice (inhalation studies) Natl. Toxicol. Program Tech. Rep. Ser. 1993;434:1–389. [PubMed] [Google Scholar]
- Neumann HG. Risk assessment of chemical carcinogens and thresholds. Crit. Rev. Toxicol. 2009;39:449–461. doi: 10.1080/10408440902810329. [DOI] [PubMed] [Google Scholar]
- Pogribny I, Yi P, James SJ. A sensitive new method for rapid detection of abnormal methylation patterns in global DNA and within CpG islands. Biochem. Biophys. Res. Commun. 1999;262:624–628. doi: 10.1006/bbrc.1999.1187. [DOI] [PubMed] [Google Scholar]
- Pogribny IP, Rusyn I, Beland FA. Epigenetic aspects of genotoxic and non-genotoxic hepatocarcinogenesis: studies in rodents. Environ. Mol. Mutagen. 2008;49:9–15. doi: 10.1002/em.20342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier MC. Chemical-induced DNA damage and human cancer risk. Nat. Rev. Cancer. 2004;4:630–637. doi: 10.1038/nrc1410. [DOI] [PubMed] [Google Scholar]
- Rappaport SM, Smith MT. Epidemiology. Environment and disease risks. Science. 2010;330:460–461. doi: 10.1126/science.1192603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusyn I, Gatti DM, Wiltshire T, Kleeberger SR, Threadgill DW. Toxicogenetics: population-based testing of drug and chemical safety in mouse models. Pharmacogenomics. 2010;11:1127–1136. doi: 10.2217/pgs.10.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss WM. Preparation of genomic DNA from mammalian tissue. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JA, Smith JA, Struhl K, editors. Current Protocols in Molecular Biology. New York, NY: Wiley-Interscience; 1989. pp. 2.2.1–2.2.3. [Google Scholar]
- Swenberg JA, Bordeerat NK, Boysen G, Carro S, Georgieva NI, Nakamura J, Troutman JM, Upton PB, Albertini RJ, Vacek PM, et al. 1,3-Butadiene: biomarkers and application to risk assessment. Chem. Biol. Interact. 2010 doi: 10.1016/j.cbi.2010.10.010. Advance Access published on October 23, 2010; doi:10.1016/j.cbi.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swenberg JA, Ham AJ, Koc H, Morinello E, Ranasinghe A, Tretyakova N, Upton PB, Wu KY. DNA adducts: effects of low exposure to ethylene oxide, vinyl chloride and butadiene. Mutat. Res. 2000;464:77–86. doi: 10.1016/s1383-5718(99)00168-0. [DOI] [PubMed] [Google Scholar]
- Tryndyak VP, Muskhelishvili L, Kovalchuk O, Rodriguez-Juarez R, Montgomery B, Churchwell MI, Ross SA, Beland FA, Pogribny IP. Effect of long-term tamoxifen exposure on genotoxic and epigenetic changes in rat liver: implications for tamoxifen-induced hepatocarcinogenesis. Carcinogenesis. 2006;27:1713–1720. doi: 10.1093/carcin/bgl050. [DOI] [PubMed] [Google Scholar]
- Wild CP. Environmental exposure measurement in cancer epidemiology. Mutagenesis. 2009;24:117–125. doi: 10.1093/mutage/gen061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Wang JR, Didion JP, Buus RJ, Bell TA, Welsh CE, Bonhomme F, Boursot P, Harr B, Yu AHT, et al. Genome-wide maps of subspecific origin and identity by descent in the laboratory mouse. Nat. Genet. 2011 doi: 10.1038/ng.847. Advance Access published on May 29, 2011; doi:10.1038/ng.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler J. Health risk assessment research: the OTA report. Environ. Health Perspect. 1993;101:402–406. doi: 10.1289/ehp.101-1519854. [DOI] [PMC free article] [PubMed] [Google Scholar]