Adipocyte associated glucocorticoid signaling regulates normal fibroblast function which is lost in inflammatory arthritis
- PMID: 39543086
- PMCID: PMC11564742
- DOI: 10.1038/s41467-024-52586-x
Adipocyte associated glucocorticoid signaling regulates normal fibroblast function which is lost in inflammatory arthritis
Abstract
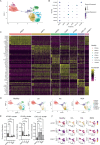
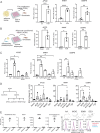
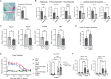
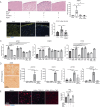
Fibroblasts play critical roles in tissue homeostasis, but in pathologic states they can drive fibrosis, inflammation, and tissue destruction. Little is known about what regulates the homeostatic functions of fibroblasts. Here, we perform RNA sequencing and identify a gene expression program in healthy synovial fibroblasts characterized by enhanced fatty acid metabolism and lipid transport. We identify cortisol as the key driver of the healthy fibroblast phenotype and that depletion of adipocytes, which express high levels of Hsd11b1, results in loss of the healthy fibroblast phenotype in mouse synovium. Additionally, fibroblast-specific glucocorticoid receptor Nr3c1 deletion in vivo leads to worsened arthritis. Cortisol signaling in fibroblasts mitigates matrix remodeling induced by TNF and TGF-β1 in vitro, while stimulation with these cytokines represses cortisol signaling and adipogenesis. Together, these findings demonstrate the importance of adipocytes and cortisol signaling in driving the healthy synovial fibroblast state that is lost in disease.
© 2024. The Author(s).
Conflict of interest statement
Figures



Update of
-
Adipocytes regulate fibroblast function, and their loss contributes to fibroblast dysfunction in inflammatory diseases.bioRxiv [Preprint]. 2023 May 18:2023.05.16.540975. doi: 10.1101/2023.05.16.540975. bioRxiv. 2023. Update in: Nat Commun. 2024 Nov 14;15(1):9859. doi: 10.1038/s41467-024-52586-x PMID: 37292637 Free PMC article. Updated. Preprint.
References
-
- Favero, M. et al. Infrapatellar fat pad features in osteoarthritis: a histopathological and molecular study. Rheumatology56, 1784–1793 (2017). - PubMed
-
- Kitagawa, T. et al. Histopathological study of the infrapatellar fat pad in the rat model of patellar tendinopathy: a basic study. Knee26, 14–19 (2019). - PubMed
MeSH terms
Substances
Grants and funding
- UH2 AR067685/AR/NIAMS NIH HHS/United States
- UM2 AR067678/AR/NIAMS NIH HHS/United States
- P30 AR070253/AR/NIAMS NIH HHS/United States
- T32 AR007530/AR/NIAMS NIH HHS/United States
- UH2 AR067681/AR/NIAMS NIH HHS/United States
- UH2 AR067688/AR/NIAMS NIH HHS/United States
- UH2 AR067689/AR/NIAMS NIH HHS/United States
- T32AR007530-37/U.S. Department of Health & Human Services | NIH | National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS)
- UH2 AR067690/AR/NIAMS NIH HHS/United States
- UH2 AR067677/AR/NIAMS NIH HHS/United States
- UH2 AR067694/AR/NIAMS NIH HHS/United States
- UH2 AR067679/AR/NIAMS NIH HHS/United States
- UH2 AR067676/AR/NIAMS NIH HHS/United States
- UH2 AR067691/AR/NIAMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical


