Mapping the future of oxidative RNA damage in neurodegeneration: Rethinking the status quo with new tools
- PMID: 39495912
- PMCID: PMC11572933
- DOI: 10.1073/pnas.2317860121
Mapping the future of oxidative RNA damage in neurodegeneration: Rethinking the status quo with new tools
Abstract
Over two decades ago, increased levels of RNA oxidation were reported in postmortem patients with ALS, Alzheimer's, Parkinson's, and other neurodegenerative diseases. Interestingly, not all cell types and transcripts were equally oxidized. Furthermore, it was shown that RNA oxidation is an early phenomenon, altogether indicating that oxidative RNA damage could be a driver, and not a consequence, of disease. Despite all these exciting observations, the field appears to have stagnated since then. We argue that this is a consequence of the shortcomings of technologies to model these diseases, limiting our understanding of which transcripts are being oxidized, which RNA-binding proteins are interacting with these RNAs, what their implications are in RNA processing, and as a result, what their potential role is in disease onset and progression. Here, we discuss the limits of previous technologies and propose ways by which advancements in iPSC-derived disease modeling, proteomics, and sequencing technologies can be combined and leveraged to answer new and decades-old questions.
Keywords: RNA-binding proteins; induced pluripotent stem cells; neurodegeneration; oxidative RNA damage; oxidative stress.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures
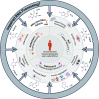

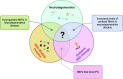
Similar articles
-
Role of RNA Oxidation in Neurodegenerative Diseases.Int J Mol Sci. 2020 Jul 16;21(14):5022. doi: 10.3390/ijms21145022. Int J Mol Sci. 2020. PMID: 32708667 Free PMC article. Review.
-
Oxidative damage to RNA in aging and neurodegenerative disorders.Neurotox Res. 2012 Oct;22(3):231-48. doi: 10.1007/s12640-012-9331-x. Epub 2012 Jun 6. Neurotox Res. 2012. PMID: 22669748 Review.
-
[Role of oxidative RNA damage in aging and neurodegenerative disorders].Brain Nerve. 2013 Feb;65(2):179-94. Brain Nerve. 2013. PMID: 23399675 Review. Japanese.
-
Stress-Specific Spatiotemporal Responses of RNA-Binding Proteins in Human Stem-Cell-Derived Motor Neurons.Int J Mol Sci. 2020 Nov 6;21(21):8346. doi: 10.3390/ijms21218346. Int J Mol Sci. 2020. PMID: 33172210 Free PMC article.
-
RNA processing-associated molecular mechanisms of neurodegenerative diseases.J Appl Genet. 2016 Aug;57(3):323-33. doi: 10.1007/s13353-015-0330-5. Epub 2015 Dec 3. J Appl Genet. 2016. PMID: 26634851 Review.
Cited by
-
RNA dynamics in oxidative stress: From obscurity to mechanistic understanding in health and disease.Proc Natl Acad Sci U S A. 2024 Nov 12;121(46):e2420445121. doi: 10.1073/pnas.2420445121. Epub 2024 Nov 4. Proc Natl Acad Sci U S A. 2024. PMID: 39495921 Free PMC article. No abstract available.
References
-
- Wilson D. M., et al. , Hallmarks of neurodegenerative diseases. Cell 186, 693–714 (2023). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous


