Nitrate-dependent antimony oxidase in an uncultured Symbiobacteriaceae member
- PMID: 39413245
- PMCID: PMC11521347
- DOI: 10.1093/ismejo/wrae204
Nitrate-dependent antimony oxidase in an uncultured Symbiobacteriaceae member
Abstract
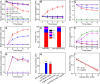
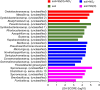
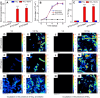
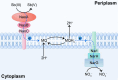
Autotrophic antimony (Sb) oxidation coupled to nitrate reduction plays an important role in the transformation and detoxification of Sb. However, the specific oxidase involved in this process has yet to be identified. Herein, we enriched the microbiota capable of nitrate-dependent Sb(III) oxidation and identified a new Sb(III) oxidase in an uncultured member of Symbiobacteriaceae. Incubation experiments demonstrated that nitrate-dependent Sb(III) oxidation occurred in the microcosm supplemented with Sb(III) and nitrate. Both the 16S rRNA gene and metagenomic analyses indicated that a species within Symbiobacteriaceae played a crucial role in this process. Furthermore, carbon-13 isotope labeling with carbon dioxide-fixing Rhodopseudomonas palustris in combination with nanoscale secondary ion mass spectrometry revealed that a newly characterized oxidase from the dimethylsulfoxide reductase family, designated as NaoABC, was responsible for autotrophic Sb(III) oxidation coupled with nitrate reduction. The NaoABC complex functions in conjunction with the nitrate reductase NarGHI, forming a redox loop that transfers electrons from Sb(III) to nitrate, thereby generating the energy necessary for autotrophic growth. This research offers new insights into the understanding of how microbes link Sb and nitrogen biogeochemical cycles in the environment.
Keywords: Symbiobacteriaceae; antimony oxidase; nitrate-dependent Sb(III) oxidation.
© The Author(s) 2024. Published by Oxford University Press on behalf of the International Society for Microbial Ecology.
Conflict of interest statement
None declared.
Figures


Similar articles
-
Microbiological oxidation of antimony(III) with oxygen or nitrate by bacteria isolated from contaminated mine sediments.Appl Environ Microbiol. 2015 Dec;81(24):8478-88. doi: 10.1128/AEM.01970-15. Epub 2015 Oct 2. Appl Environ Microbiol. 2015. PMID: 26431974 Free PMC article.
-
Influence of the Chemical Form of Antimony on Soil Microbial Community Structure and Arsenite Oxidation Activity.Microbes Environ. 2018 Jul 4;33(2):214-221. doi: 10.1264/jsme2.ME17182. Epub 2018 Jun 9. Microbes Environ. 2018. PMID: 29887548 Free PMC article.
-
Identification of antimony- and arsenic-oxidizing bacteria associated with antimony mine tailing.Microbes Environ. 2013;28(2):257-63. doi: 10.1264/jsme2.me12217. Epub 2013 May 11. Microbes Environ. 2013. PMID: 23666539 Free PMC article.
-
Enhanced biological antimony removal from water by combining elemental sulfur autotrophic reduction and disproportionation.J Hazard Mater. 2022 Jul 15;434:128926. doi: 10.1016/j.jhazmat.2022.128926. Epub 2022 Apr 14. J Hazard Mater. 2022. PMID: 35452992
-
Sulfur-driven autotrophic denitrification: diversity, biochemistry, and engineering applications.Appl Microbiol Biotechnol. 2010 Nov;88(5):1027-42. doi: 10.1007/s00253-010-2847-1. Epub 2010 Sep 1. Appl Microbiol Biotechnol. 2010. PMID: 20809074 Review.
References
-
- Zhang Y, Ding C, Gong Det al. . A review of the environmental chemical behavior, detection and treatment of antimony. Environ Technol Innov 2021;24:102026. 10.1016/j.eti.2021.102026 - DOI
-
- Tschan M, Robinson B, Johnson CAet al. . Antimony uptake and toxicity in sunflower and maize growing in Sb(III) and Sb(V) contaminated soil. Plant Soil 2010;334:235–45. 10.1007/s11104-010-0378-2 - DOI
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources


