This is a preprint.
A Phylogenetic Model of Established and Enabled Biome Shifts
- PMID: 39282335
- PMCID: PMC11398350
- DOI: 10.1101/2024.08.30.610561
A Phylogenetic Model of Established and Enabled Biome Shifts
Abstract
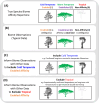
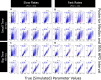
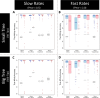
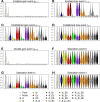
Where each species actually lives is distinct from where it could potentially survive and persist. This suggests that it may be important to distinguish established from enabled biome affinities when considering how ancestral species moved and evolved among major habitat types. We introduce a new phylogenetic method, called RFBS, to model how anagenetic and cladogenetic events cause established and enabled biome affinities (or, more generally, other discrete realized versus fundamental niche states) to shift over evolutionary timescale. We provide practical guidelines for how to assign established and enabled biome affinity states to extant taxa, using the flowering plant clade Viburnum as a case study. Through a battery of simulation experiments, we show that RFBS performs well, even when we have realistically imperfect knowledge of enabled biome affinities for most analyzed species. We also show that RFBS reliably discerns established from enabled affinities, with similar accuracy to standard competing models that ignore the existence of enabled biome affinities. Lastly, we apply RFBS to Viburnum to infer ancestral biomes throughout the tree and to highlight instances where repeated shifts between established affinities for warm and cold temperate forest biomes were enabled by a stable and slowly-evolving enabled affinity for both temperate biomes.
Keywords: Bayesian inference; Viburnum; biome shift; niche evolution; phylogenetic model.
Figures


Similar articles
-
Joint Phylogenetic Estimation of Geographic Movements and Biome Shifts during the Global Diversification of Viburnum.Syst Biol. 2021 Jan 1;70(1):67-85. doi: 10.1093/sysbio/syaa027. Syst Biol. 2021. PMID: 32267945
-
Modeling Phylogenetic Biome Shifts on a Planet with a Past.Syst Biol. 2021 Jan 1;70(1):86-107. doi: 10.1093/sysbio/syaa045. Syst Biol. 2021. PMID: 32514540
-
Diversification and trait evolution in New Zealand woody lineages across changing biomes.J R Soc N Z. 2022 Aug 17;54(1):98-123. doi: 10.1080/03036758.2022.2108071. eCollection 2024. J R Soc N Z. 2022. PMID: 39439477 Free PMC article.
-
Birth of a biome: insights into the assembly and maintenance of the Australian arid zone biota.Mol Ecol. 2008 Oct;17(20):4398-417. doi: 10.1111/j.1365-294X.2008.03899.x. Epub 2008 Aug 27. Mol Ecol. 2008. PMID: 18761619 Review.
-
Alternative Biome States in Terrestrial Ecosystems.Trends Plant Sci. 2020 Mar;25(3):250-263. doi: 10.1016/j.tplants.2019.11.003. Epub 2020 Jan 6. Trends Plant Sci. 2020. PMID: 31917105 Review.
References
-
- Anderson Eric C., Ng Thomas C., Crandall Eric D., and Garza John Carlos. Genetic and individual assignment of tetraploid green sturgeon with SNP assay data. Conservation Genetics, 18(5):1119–1130, October 2017. ISSN 1566–0621, 1572–9737. doi: 10.1007/s10592-017-0963-5. URL 10.1007/s10592-017-0963-5. - DOI - DOI
-
- Antonelli Alexandre, Zizka Alexander, Carvalho Fernanda Antunes, Scharn Ruud, Bacon Christine D., Silvestro Daniele, and Condamine Fabien L.. Amazonia is the primary source of Neotropical biodiversity. Proceedings of the National Academy of Sciences, 115(23):6034–6039, June 2018. ISSN 0027–8424, 1091–6490. doi: 10.1073/pnas.1713819115. URL 10.1073/pnas.1713819115. - DOI - DOI - PMC - PubMed
-
- Araújo Miguel B. and Luoto Miska. The importance of biotic interactions for modelling species distributions under climate change. Global Ecology and Biogeography, 16(6):743–753, 2007. ISSN 1466–8238. doi: 10.1111/j.1466-8238.2007.00359.x. URL https://onlinelibrary.wiley.com/doi/abs/10.1111/j.1466-8238.2007.00359.x. _eprint: https://onlinelibrary.wiley.com/doi/pdf/10.1111/j.1466-8238.2007.00359.x. - DOI - DOI - DOI
-
- Axelrod Daniel I.. Edaphic Aridity as a Factor in Angiosperm Evolution. The American Naturalist, 106(949):311–320, 1972. ISSN 0003–0147. URL https://www.jstor.org/stable/2459779. Publisher: [University of Chicago Press, American Society of Naturalists; ].
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

