Comparative Population Genomics of Arctic Sled Dogs Reveals a Deep and Complex History
- PMID: 39193769
- PMCID: PMC11403282
- DOI: 10.1093/gbe/evae190
Comparative Population Genomics of Arctic Sled Dogs Reveals a Deep and Complex History
Abstract
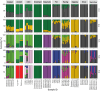
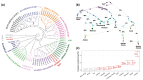
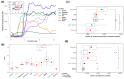
Recent evidence demonstrates genomic and morphological continuity in the Arctic ancestral lineage of dogs. Here, we use the Siberian Husky to investigate the genomic legacy of the northeast Eurasian Arctic lineage and model the deep population history using genome-wide single nucleotide polymorphisms. Utilizing ancient dog-calibrated molecular clocks, we found that at least two distinct lineages of Arctic dogs existed in ancient Eurasia at the end of the Pleistocene. This pushes back the origin of sled dogs in the northeast Siberian Arctic with humans likely intentionally selecting dogs to perform different functions and keeping breeding populations that overlap in time and space relatively reproductively isolated. In modern Siberian Huskies, we found significant population structure based on how they are used by humans, recent European breed introgression in about half of the dogs that participate in races, moderate levels of inbreeding, and fewer potentially harmful variants in populations under strong selection for form and function (show, sled show, and racing populations of Siberian Huskies). As the struggle to preserve unique evolutionary lineages while maintaining genetic health intensifies across pedigreed dogs, understanding the genomic history to guide policies and best practices for breed management is crucial to sustain these ancient lineages and their unique evolutionary identity.
Keywords: Arctic dogs; Siberian Husky; canine evolution; canine genetic health; sled dogs.
© The Author(s) 2024. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Conflict of interest statement
T.A.S. has both show and racing Siberian Huskies and participates in AKC conformation and sled dog racing sports. H.J.H. has formerly participated in sled dog races with Alaskan sled dogs. K.S. declares no competing interests.
Figures



Similar articles
-
Description of breed ancestry and genetic health traits in arctic sled dog breeds.Canine Med Genet. 2021 Sep 20;8(1):8. doi: 10.1186/s40575-021-00108-z. Canine Med Genet. 2021. PMID: 34544496 Free PMC article.
-
Arctic-adapted dogs emerged at the Pleistocene-Holocene transition.Science. 2020 Jun 26;368(6498):1495-1499. doi: 10.1126/science.aaz8599. Science. 2020. PMID: 32587022 Free PMC article.
-
Using multiple markers to elucidate the ancient, historical and modern relationships among North American Arctic dog breeds.Heredity (Edinb). 2015 Dec;115(6):488-95. doi: 10.1038/hdy.2015.49. Epub 2015 Jun 24. Heredity (Edinb). 2015. PMID: 26103948 Free PMC article.
-
Deciphering the Origin of Dogs: From Fossils to Genomes.Annu Rev Anim Biosci. 2017 Feb 8;5:281-307. doi: 10.1146/annurev-animal-022114-110937. Epub 2016 Nov 28. Annu Rev Anim Biosci. 2017. PMID: 27912242 Review.
-
Cystic echinococcosis in the Arctic and Sub-Arctic.Parasitology. 2003;127 Suppl:S73-85. doi: 10.1017/s0031182003003664. Parasitology. 2003. PMID: 15027606 Review.
References
-
- Ameen C, Feuerborn TR, Brown SK, Linderholm A, Hulme-Beaman A, Lebrasseur O, Sinding MHS, Lounsberry ZT, Lin AT, Appelt M, et al. Specialized sledge dogs accompanied Inuit dispersal across the North American Arctic. Proc Biol Sci. 2019:286(1916):20191929. 10.1098/rspb.2019.1929. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials


