Essential and multifunctional mpox virus E5 helicase-primase in double and single hexamer
- PMID: 39167653
- PMCID: PMC11338233
- DOI: 10.1126/sciadv.adl1150
Essential and multifunctional mpox virus E5 helicase-primase in double and single hexamer
Abstract
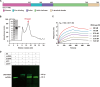
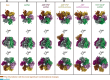
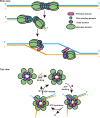
An outbreak of mpox virus in May 2022 has spread over 110 nonpandemic regions in the world, posing a great threat to global health. Mpox virus E5, a helicase-primase, plays an essential role in DNA replication, but the molecular mechanisms are elusive. Here, we report seven structures of mpox virus E5 in a double hexamer (DH) and six in single hexamer in different conformations, indicating a rotation mechanism for helicase and a coupling action for primase. The DH is formed through the interface of zinc-binding domains, and the central channel density indicates potential double-stranded DNA (dsDNA), which helps to identify dsDNA binding residues Arg249, Lys286, Lys315, and Lys317. Our work is important not only for understanding poxviral DNA replication but also for the development of novel therapeutics for serious poxviral infections including smallpox virus and mpox virus.
Figures


Similar articles
-
Domain Organization of Vaccinia Virus Helicase-Primase D5.J Virol. 2016 Apr 14;90(9):4604-4613. doi: 10.1128/JVI.00044-16. Print 2016 May. J Virol. 2016. PMID: 26912611 Free PMC article.
-
An in trans interaction at the interface of the helicase and primase domains of the hexameric gene 4 protein of bacteriophage T7 modulates their activities.J Biol Chem. 2009 Aug 28;284(35):23842-51. doi: 10.1074/jbc.M109.026104. Epub 2009 Jul 1. J Biol Chem. 2009. PMID: 19574219 Free PMC article.
-
Dynamic coupling between the motors of DNA replication: hexameric helicase, DNA polymerase, and primase.Curr Opin Chem Biol. 2011 Oct;15(5):595-605. doi: 10.1016/j.cbpa.2011.08.003. Epub 2011 Aug 22. Curr Opin Chem Biol. 2011. PMID: 21865075 Free PMC article. Review.
-
Structural insight into the assembly and working mechanism of helicase-primase D5 from Mpox virus.Nat Struct Mol Biol. 2024 Jan;31(1):68-81. doi: 10.1038/s41594-023-01142-0. Epub 2024 Jan 4. Nat Struct Mol Biol. 2024. PMID: 38177671
-
Choreography of bacteriophage T7 DNA replication.Curr Opin Chem Biol. 2011 Oct;15(5):580-6. doi: 10.1016/j.cbpa.2011.07.024. Epub 2011 Sep 9. Curr Opin Chem Biol. 2011. PMID: 21907611 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources


