Neurogenesis in Caenorhabditis elegans
- PMID: 39167071
- PMCID: PMC11457946
- DOI: 10.1093/genetics/iyae116
Neurogenesis in Caenorhabditis elegans
Abstract
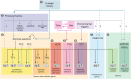
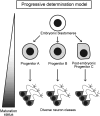
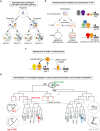
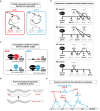
Animals rely on their nervous systems to process sensory inputs, integrate these with internal signals, and produce behavioral outputs. This is enabled by the highly specialized morphologies and functions of neurons. Neuronal cells share multiple structural and physiological features, but they also come in a large diversity of types or classes that give the nervous system its broad range of functions and plasticity. This diversity, first recognized over a century ago, spurred classification efforts based on morphology, function, and molecular criteria. Caenorhabditis elegans, with its precisely mapped nervous system at the anatomical level, an extensive molecular description of most of its neurons, and its genetic amenability, has been a prime model for understanding how neurons develop and diversify at a mechanistic level. Here, we review the gene regulatory mechanisms driving neurogenesis and the diversification of neuron classes and subclasses in C. elegans. We discuss our current understanding of the specification of neuronal progenitors and their differentiation in terms of the transcription factors involved and ensuing changes in gene expression and chromatin landscape. The central theme that has emerged is that the identity of a neuron is defined by modules of gene batteries that are under control of parallel yet interconnected regulatory mechanisms. We focus on how, to achieve these terminal identities, cells integrate information along their developmental lineages. Moreover, we discuss how neurons are diversified postembryonically in a time-, genetic sex-, and activity-dependent manner. Finally, we discuss how the understanding of neuronal development can provide insights into the evolution of neuronal diversity.
Keywords: Caenorhabditis elegans; WormBook; gene regulation; neurodevelopment; neurogenesis; transcription factors.
© The Author(s) 2024. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflicts of interest The authors declare no conflict of interest.
Figures



Similar articles
-
A map of terminal regulators of neuronal identity in Caenorhabditis elegans.Wiley Interdiscip Rev Dev Biol. 2016 Jul;5(4):474-98. doi: 10.1002/wdev.233. Epub 2016 May 2. Wiley Interdiscip Rev Dev Biol. 2016. PMID: 27136279 Free PMC article. Review.
-
Cis-regulatory mechanisms of left/right asymmetric neuron-subtype specification in C. elegans.Development. 2009 Jan;136(1):147-60. doi: 10.1242/dev.030064. Development. 2009. PMID: 19060335 Free PMC article.
-
Terminal Selectors of Neuronal Identity.Curr Top Dev Biol. 2016;116:455-75. doi: 10.1016/bs.ctdb.2015.12.007. Epub 2016 Jan 14. Curr Top Dev Biol. 2016. PMID: 26970634 Review.
-
In silico analysis of the transcriptional regulatory logic of neuronal identity specification throughout the C. elegans nervous system.Elife. 2021 Jun 24;10:e64906. doi: 10.7554/eLife.64906. Elife. 2021. PMID: 34165430 Free PMC article.
-
C. elegans SoxB genes are dispensable for embryonic neurogenesis but required for terminal differentiation of specific neuron types.Development. 2015 Jul 15;142(14):2464-77. doi: 10.1242/dev.125740. Epub 2015 Jul 7. Development. 2015. PMID: 26153233 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources


