CrysFormer: Protein structure determination via Patterson maps, deep learning, and partial structure attention
- PMID: 39148510
- PMCID: PMC11326852
- DOI: 10.1063/4.0000252
CrysFormer: Protein structure determination via Patterson maps, deep learning, and partial structure attention
Abstract
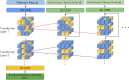
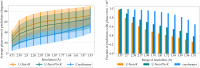
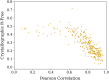
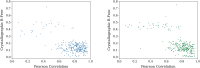
Determining the atomic-level structure of a protein has been a decades-long challenge. However, recent advances in transformers and related neural network architectures have enabled researchers to significantly improve solutions to this problem. These methods use large datasets of sequence information and corresponding known protein template structures, if available. Yet, such methods only focus on sequence information. Other available prior knowledge could also be utilized, such as constructs derived from x-ray crystallography experiments and the known structures of the most common conformations of amino acid residues, which we refer to as partial structures. To the best of our knowledge, we propose the first transformer-based model that directly utilizes experimental protein crystallographic data and partial structure information to calculate electron density maps of proteins. In particular, we use Patterson maps, which can be directly obtained from x-ray crystallography experimental data, thus bypassing the well-known crystallographic phase problem. We demonstrate that our method, CrysFormer, achieves precise predictions on two synthetic datasets of peptide fragments in crystalline forms, one with two residues per unit cell and the other with fifteen. These predictions can then be used to generate accurate atomic models using established crystallographic refinement programs.
© 2024 Author(s).
Conflict of interest statement
The authors have no conflicts to disclose.
Figures





Similar articles
-
A deep learning solution for crystallographic structure determination.IUCrJ. 2023 Jul 1;10(Pt 4):487-496. doi: 10.1107/S2052252523004293. IUCrJ. 2023. PMID: 37409806 Free PMC article.
-
Determining the Crystal Structure of TRPV6.In: Kozak JA, Putney JW Jr, editors. Calcium Entry Channels in Non-Excitable Cells. Boca Raton (FL): CRC Press/Taylor & Francis; 2018. Chapter 14. In: Kozak JA, Putney JW Jr, editors. Calcium Entry Channels in Non-Excitable Cells. Boca Raton (FL): CRC Press/Taylor & Francis; 2018. Chapter 14. PMID: 30299652 Free Books & Documents. Review.
-
A flexible and efficient procedure for the solution and phase refinement of protein structures.Acta Crystallogr D Biol Crystallogr. 2000 Sep;56(Pt 9):1137-47. doi: 10.1107/s090744490000932x. Acta Crystallogr D Biol Crystallogr. 2000. PMID: 10957632
-
Ab initio and template-based prediction of multi-class distance maps by two-dimensional recursive neural networks.BMC Struct Biol. 2009 Jan 30;9:5. doi: 10.1186/1472-6807-9-5. BMC Struct Biol. 2009. PMID: 19183478 Free PMC article.
-
Refinement of Atomic Structures Against cryo-EM Maps.Methods Enzymol. 2016;579:277-305. doi: 10.1016/bs.mie.2016.05.033. Epub 2016 Jun 24. Methods Enzymol. 2016. PMID: 27572731 Review.
References
-
- Tanford C. and Reynolds J., Nature's Robots: A History of Proteins ( Oxford University Press, 2004).
-
- Drenth J., Principles of Protein X-Ray Crystallography ( Springer Science & Business Media, 2007).
Grants and funding
LinkOut - more resources
Full Text Sources


