Multi-omic analysis of Huntington's disease reveals a compensatory astrocyte state
- PMID: 39112488
- PMCID: PMC11306246
- DOI: 10.1038/s41467-024-50626-0
Multi-omic analysis of Huntington's disease reveals a compensatory astrocyte state
Abstract
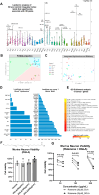
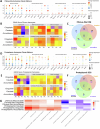
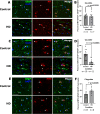
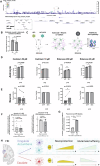
The mechanisms underlying the selective regional vulnerability to neurodegeneration in Huntington's disease (HD) have not been fully defined. To explore the role of astrocytes in this phenomenon, we used single-nucleus and bulk RNAseq, lipidomics, HTT gene CAG repeat-length measurements, and multiplexed immunofluorescence on HD and control post-mortem brains. We identified genes that correlated with CAG repeat length, which were enriched in astrocyte genes, and lipidomic signatures that implicated poly-unsaturated fatty acids in sensitizing neurons to cell death. Because astrocytes play essential roles in lipid metabolism, we explored the heterogeneity of astrocytic states in both protoplasmic and fibrous-like (CD44+) astrocytes. Significantly, one protoplasmic astrocyte state showed high levels of metallothioneins and was correlated with the selective vulnerability of distinct striatal neuronal populations. When modeled in vitro, this state improved the viability of HD-patient-derived spiny projection neurons. Our findings uncover key roles of astrocytic states in protecting against neurodegeneration in HD.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Update of
-
Multi-OMIC analysis of Huntington disease reveals a neuroprotective astrocyte state.bioRxiv [Preprint]. 2023 Sep 12:2023.09.08.556867. doi: 10.1101/2023.09.08.556867. bioRxiv. 2023. Update in: Nat Commun. 2024 Aug 8;15(1):6742. doi: 10.1038/s41467-024-50626-0 PMID: 37745577 Free PMC article. Updated. Preprint.
Similar articles
-
Multi-OMIC analysis of Huntington disease reveals a neuroprotective astrocyte state.bioRxiv [Preprint]. 2023 Sep 12:2023.09.08.556867. doi: 10.1101/2023.09.08.556867. bioRxiv. 2023. Update in: Nat Commun. 2024 Aug 8;15(1):6742. doi: 10.1038/s41467-024-50626-0 PMID: 37745577 Free PMC article. Updated. Preprint.
-
Downregulation of glial genes involved in synaptic function mitigates Huntington's disease pathogenesis.Elife. 2021 Apr 19;10:e64564. doi: 10.7554/eLife.64564. Elife. 2021. PMID: 33871358 Free PMC article.
-
Neuronal and astrocytic contributions to Huntington's disease dissected with zinc finger protein transcriptional repressors.Cell Rep. 2023 Jan 31;42(1):111953. doi: 10.1016/j.celrep.2022.111953. Epub 2023 Jan 7. Cell Rep. 2023. PMID: 36640336 Free PMC article.
-
Cell-Autonomous and Non-cell-Autonomous Pathogenic Mechanisms in Huntington's Disease: Insights from In Vitro and In Vivo Models.Neurotherapeutics. 2019 Oct;16(4):957-978. doi: 10.1007/s13311-019-00782-9. Neurotherapeutics. 2019. PMID: 31529216 Free PMC article. Review.
-
Multiple clinical features of Huntington's disease correlate with mutant HTT gene CAG repeat lengths and neurodegeneration.J Neurol. 2019 Mar;266(3):551-564. doi: 10.1007/s00415-018-8940-6. Epub 2018 Jun 28. J Neurol. 2019. PMID: 29956026 Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous


