Artificial intelligence in fusion protein three-dimensional structure prediction: Review and perspective
- PMID: 39090739
- PMCID: PMC11294035
- DOI: 10.1002/ctm2.1789
Artificial intelligence in fusion protein three-dimensional structure prediction: Review and perspective
Abstract
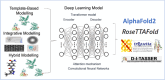
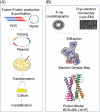
Recent advancements in artificial intelligence (AI) have accelerated the prediction of unknown protein structures. However, accurately predicting the three-dimensional (3D) structures of fusion proteins remains a difficult task because the current AI-based protein structure predictions are focused on the WT proteins rather than on the newly fused proteins in nature. Following the central dogma of biology, fusion proteins are translated from fusion transcripts, which are made by transcribing the fusion genes between two different loci through the chromosomal rearrangements in cancer. Accurately predicting the 3D structures of fusion proteins is important for understanding the functional roles and mechanisms of action of new chimeric proteins. However, predicting their 3D structure using a template-based model is challenging because known template structures are often unavailable in databases. Deep learning (DL) models that utilize multi-level protein information have revolutionized the prediction of protein 3D structures. In this review paper, we highlighted the latest advancements and ongoing challenges in predicting the 3D structure of fusion proteins using DL models. We aim to explore both the advantages and challenges of employing AlphaFold2, RoseTTAFold, tr-Rosetta and D-I-TASSER for modelling the 3D structures. HIGHLIGHTS: This review provides the overall pipeline and landscape of the prediction of the 3D structure of fusion protein. This review provides the factors that should be considered in predicting the 3D structures of fusion proteins using AI approaches in each step. This review highlights the latest advancements and ongoing challenges in predicting the 3D structure of fusion proteins using deep learning models. This review explores the advantages and challenges of employing AlphaFold2, RoseTTAFold, tr-Rosetta, and D-I-TASSER to model 3D structures.
Keywords: AI; AlphaFold2; RoseTTAFold; deep learning; fusion protein structure; protein structure prediction.
© 2024 The Author(s). Clinical and Translational Medicine published by John Wiley & Sons Australia, Ltd on behalf of Shanghai Institute of Clinical Bioinformatics.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





Similar articles
-
AI-Driven Deep Learning Techniques in Protein Structure Prediction.Int J Mol Sci. 2024 Aug 1;25(15):8426. doi: 10.3390/ijms25158426. Int J Mol Sci. 2024. PMID: 39125995 Free PMC article. Review.
-
Recent Progress of Protein Tertiary Structure Prediction.Molecules. 2024 Feb 13;29(4):832. doi: 10.3390/molecules29040832. Molecules. 2024. PMID: 38398585 Free PMC article. Review.
-
Overview of AlphaFold2 and breakthroughs in overcoming its limitations.Comput Biol Med. 2024 Jun;176:108620. doi: 10.1016/j.compbiomed.2024.108620. Epub 2024 May 15. Comput Biol Med. 2024. PMID: 38761500 Review.
-
Prediction of protein structure and AI.J Hum Genet. 2024 Oct;69(10):477-480. doi: 10.1038/s10038-023-01215-4. Epub 2024 Jan 4. J Hum Genet. 2024. PMID: 38177398 Review.
-
Review and Comparative Analysis of Methods and Advancements in Predicting Protein Complex Structure.Interdiscip Sci. 2024 Jun;16(2):261-288. doi: 10.1007/s12539-024-00626-x. Epub 2024 Jul 2. Interdiscip Sci. 2024. PMID: 38955920 Review.
Cited by
-
Advances in Computational Intelligence-Based Methods of Structure and Function Prediction of Proteins.Biomolecules. 2024 Aug 29;14(9):1083. doi: 10.3390/biom14091083. Biomolecules. 2024. PMID: 39334850 Free PMC article.
References
-
- Kuhlman B, Dantas G, Ireton GC, Varani G, Stoddard BL, Baker D. Design of a novel globular protein fold with atomic‐level accuracy. Science. 2003;302:1364‐1368. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

