Plasmid DNA Complexes in Powder Form Studied by Spectroscopic and Diffraction Methods
- PMID: 39063822
- PMCID: PMC11278597
- DOI: 10.3390/ma17143530
Plasmid DNA Complexes in Powder Form Studied by Spectroscopic and Diffraction Methods
Abstract
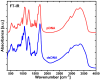
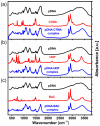
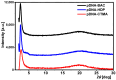
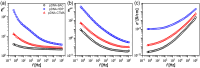
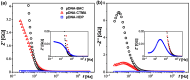
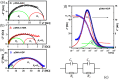
Currently, new functional materials are being created with a strong emphasis on their ecological aspect. Materials and devices based on DNA biopolymers, being environmentally friendly, are therefore very interesting from the point of view of applications. In this paper, we present the results of research on complexes in the powder form based on plasmid DNA (pDNA) and three surfactants with aliphatic chains containing 16 carbon atoms (cetyltrimethylammonium chloride, benzyldimethylhexadecylammonium chloride and hexadecylpyridinium chloride). The X-ray diffraction results indicate a local hexagonal packing of DNA helices in plasmid DNA complexes, resembling the packing for corresponding complexes based on linear DNA. Based on the Fourier-transform infrared spectroscopy results, the DNA conformation in all three complexes was determined as predominantly of A-type. The two relaxation processes revealed by dielectric spectroscopy for all the studied complexes are connected with two different contributions to total conductivity (crystallite part and grain boundaries). The crystallite part (grain interior) was interpreted as an oscillation of the polar surfactant head groups and is dependent on the conformation of the surfactant chain. The influence of the DNA type on the properties of the complexes is discussed, taking into account our previous results for complexes based on linear DNA. We showed that the type of DNA has an impact on the properties of the complexes, which has not been demonstrated so far. It was also found that the layer of pDNA-surfactant complexes can be used as a layer with variable specific electric conductivity by selecting the frequency, which is interesting from an application point of view.
Keywords: DNA complex; X-ray diffraction; dielectric spectroscopy; infrared spectroscopy; plasmid DNA.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Effect of double-tailed surfactant architecture on the conformation, self-assembly, and processing in polypeptide-surfactant complexes.Biomacromolecules. 2009 Oct 12;10(10):2787-94. doi: 10.1021/bm900630u. Biomacromolecules. 2009. PMID: 19645442
-
The nanostructure of surfactant-DNA complexes with different arrangements.Colloids Surf B Biointerfaces. 2013 Nov 1;111:663-71. doi: 10.1016/j.colsurfb.2013.06.057. Epub 2013 Jul 10. Colloids Surf B Biointerfaces. 2013. PMID: 23907055
-
Molecular mechanism and thermodynamics study of plasmid DNA and cationic surfactants interactions.Langmuir. 2006 Apr 11;22(8):3735-43. doi: 10.1021/la052161s. Langmuir. 2006. PMID: 16584250
-
The physical properties of glycosyl diacylglycerols. Calorimetric, X-ray diffraction and Fourier transform spectroscopic studies of a homologous series of 1,2-di-O-acyl-3-O-(beta-D-galactopyranosyl)-sn-glycerols.Chem Phys Lipids. 2001 Jun;111(2):139-61. doi: 10.1016/s0009-3084(01)00153-0. Chem Phys Lipids. 2001. PMID: 11457442
-
DNA-surfactant complexes: self-assembly properties and applications.Chem Soc Rev. 2017 Aug 14;46(16):5147-5172. doi: 10.1039/c7cs00165g. Chem Soc Rev. 2017. PMID: 28686247 Review.
References
-
- Cooper G.M., Hausman R.E. The Cell: A Molecular Approach. 6th ed. Volume 4 Sinauer Associates; Sunderland, MA, USA: 2007.
-
- Mielke C., Benham S. DNA mechanics. Annu. Rev. Biomed. Eng. 2005;7:21. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources


