Trait-mediated speciation and human-driven extinctions in proboscideans revealed by unsupervised Bayesian neural networks
- PMID: 39047110
- PMCID: PMC11268411
- DOI: 10.1126/sciadv.adl2643
Trait-mediated speciation and human-driven extinctions in proboscideans revealed by unsupervised Bayesian neural networks
Abstract
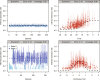
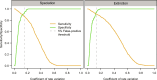
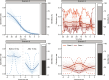
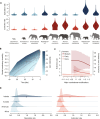
Species life-history traits, paleoenvironment, and biotic interactions likely influence speciation and extinction rates, affecting species richness over time. Birth-death models inferring the impact of these factors typically assume monotonic relationships between single predictors and rates, limiting our ability to assess more complex effects and their relative importance and interaction. We introduce a Bayesian birth-death model using unsupervised neural networks to explore multifactorial and nonlinear effects on speciation and extinction rates using fossil data. It infers lineage- and time-specific rates and disentangles predictor effects and importance through explainable artificial intelligence techniques. Analysis of the proboscidean fossil record revealed speciation rates shaped by dietary flexibility and biogeographic events. The emergence of modern humans escalated extinction rates, causing recent diversity decline, while regional climate had a lesser impact. Our model paves the way for an improved understanding of the intricate dynamics shaping clade diversification.
Figures
Similar articles
-
Bayesian estimation of speciation and extinction from incomplete fossil occurrence data.Syst Biol. 2014 May;63(3):349-67. doi: 10.1093/sysbio/syu006. Epub 2014 Feb 8. Syst Biol. 2014. PMID: 24510972 Free PMC article.
-
Heritability of extinction rates links diversification patterns in molecular phylogenies and fossils.Syst Biol. 2009 Dec;58(6):629-40. doi: 10.1093/sysbio/syp069. Epub 2009 Oct 5. Syst Biol. 2009. PMID: 20525614
-
Interactions within and between clades shaped the diversification of terrestrial carnivores.Evolution. 2017 Jul;71(7):1855-1864. doi: 10.1111/evo.13269. Epub 2017 Jun 8. Evolution. 2017. PMID: 28543226
-
Why are there so many insect species? Perspectives from fossils and phylogenies.Biol Rev Camb Philos Soc. 2007 Aug;82(3):425-54. doi: 10.1111/j.1469-185X.2007.00018.x. Biol Rev Camb Philos Soc. 2007. PMID: 17624962 Review.
-
Is regional species diversity bounded or unbounded?Biol Rev Camb Philos Soc. 2013 Feb;88(1):140-65. doi: 10.1111/j.1469-185X.2012.00245.x. Epub 2012 Sep 7. Biol Rev Camb Philos Soc. 2013. PMID: 22958676 Review.
References
-
- Blanco F., Calatayud J., Martín-Perea D. M., Domingo M. S., Menéndez I., Müller J., Fernández M. H., Cantalapiedra J. L., Punctuated ecological equilibrium in mammal communities over evolutionary time scales. Science 372, 300–303 (2021). - PubMed
-
- Fraser D., Villaseñor A., Tóth A. B., Balk M. A., Eronen J. T., Andrew Barr W., Behrensmeyer A. K., Davis M., Du A., Tyler Faith J., Graves G. R., Gotelli N. J., Jukar A. M., Looy C. V., McGill B. J., Miller J. H., Pineda-Munoz S., Potts R., Shupinski A. B., Soul L. C., Kathleen Lyons S., Late quaternary biotic homogenization of North American mammalian faunas. Nat. Commun. 13, 3940 (2022). - PMC - PubMed
-
- Rolland J., Condamine F. L., Beeravolu C. R., Jiguet F., Morlon H., Dispersal is a major driver of the latitudinal diversity gradient of Carnivora. Glob. Ecol. Biogeogr. 24, 1059–1071 (2015).
-
- Rabosky D. L., Chang J., Title P. O., Cowman P. F., Sallan L., Friedman M., Kaschner K., Garilao C., Near T. J., Coll M., Alfaro M. E., An inverse latitudinal gradient in speciation rate for marine fishes. Nature 559, 392–395 (2018). - PubMed
-
- Fenton I. S., Aze T., Farnsworth A., Valdes P., Saupe E. E., Origination of the modern-style diversity gradient 15 million years ago. Nature 614, 708–712 (2023). - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources


