Exploring the Structurally Conserved Regions and Functional Significance in Bacterial N-Terminal Nucleophile (Ntn) Amide-Hydrolases
- PMID: 38999960
- PMCID: PMC11241749
- DOI: 10.3390/ijms25136850
Exploring the Structurally Conserved Regions and Functional Significance in Bacterial N-Terminal Nucleophile (Ntn) Amide-Hydrolases
Abstract
The initial adoption of penicillin as an antibiotic marked the start of exploring other compounds essential for pharmaceuticals, yet resistance to penicillins and their side effects has compromised their efficacy. The N-terminal nucleophile (Ntn) amide-hydrolases S45 family plays a key role in catalyzing amide bond hydrolysis in various compounds, including antibiotics like penicillin and cephalosporin. This study comprehensively analyzes the structural and functional traits of the bacterial N-terminal nucleophile (Ntn) amide-hydrolases S45 family, covering penicillin G acylases, cephalosporin acylases, and D-succinylase. Utilizing structural bioinformatics tools and sequence analysis, the investigation delineates structurally conserved regions (SCRs) and substrate binding site variations among these enzymes. Notably, sixteen SCRs crucial for substrate interaction are identified solely through sequence analysis, emphasizing the significance of sequence data in characterizing functionally relevant regions. These findings introduce a novel approach for identifying targets to enhance the biocatalytic properties of N-terminal nucleophile (Ntn) amide-hydrolases, while facilitating the development of more accurate three-dimensional models, particularly for enzymes lacking structural data. Overall, this research advances our understanding of structure-function relationships in bacterial N-terminal nucleophile (Ntn) amide-hydrolases, providing insights into strategies for optimizing their enzymatic capabilities.
Keywords: N-terminal nucleophile (Ntn) amide-hydrolase; protein design; protein modeling; structure–function relationship.
Conflict of interest statement
The authors certify that they have NO affiliations with or involvement in any organization or entity with any financial interest (such as honoraria; educational grants; participation in speakers’ bureaus; membership, employment, consultancies, stock ownership, or other equity interest; and expert testimony or patent-licensing arrangements), or non-financial interest (such as personal or professional relationships, affiliations, and knowledge or beliefs) in the subject matter or materials discussed in this manuscript.
Figures



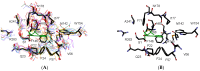
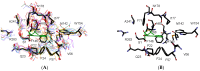
Similar articles
-
Peptidase family U34 belongs to the superfamily of N-terminal nucleophile hydrolases.Protein Sci. 2003 May;12(5):1131-5. doi: 10.1110/ps.0240803. Protein Sci. 2003. PMID: 12717035 Free PMC article.
-
The Human Ntn-Hydrolase Superfamily: Structure, Functions and Perspectives.Cells. 2022 May 10;11(10):1592. doi: 10.3390/cells11101592. Cells. 2022. PMID: 35626629 Free PMC article. Review.
-
Structural comparison of Ntn-hydrolases.Protein Sci. 2000 Dec;9(12):2329-37. doi: 10.1110/ps.9.12.2329. Protein Sci. 2000. PMID: 11206054 Free PMC article.
-
Structural and enzymatic analysis of a dimeric cholylglycine hydrolase like acylase active on N-acyl homoserine lactones.Biochimie. 2020 Oct;177:108-116. doi: 10.1016/j.biochi.2020.07.017. Epub 2020 Aug 21. Biochimie. 2020. PMID: 32835734
-
Modeling and molecular dynamics indicate that snake venom phospholipase B-like enzymes are Ntn-hydrolases.Toxicon. 2018 Oct;153:106-113. doi: 10.1016/j.toxicon.2018.08.014. Epub 2018 Sep 1. Toxicon. 2018. PMID: 30179630 Review.
References
-
- Umaru I.J., Adam B.R., Habibu B., Umaru K.I., Chizaram B.C. Biochemical Impact of Microorganism and Enzymatic Activities in Food and Pharmaceutical Industries. Int. J. Adv. Biochem. Res. 2021;5:42–57. doi: 10.33545/26174693.2021.v5.i1a.147. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials


