Pathogenic variants in human DNA damage repair genes mostly arose after the latest human out-of-Africa migration
- PMID: 38948361
- PMCID: PMC11211533
- DOI: 10.3389/fgene.2024.1408952
Pathogenic variants in human DNA damage repair genes mostly arose after the latest human out-of-Africa migration
Abstract
Introduction: The DNA damage repair (DDR) system in human genome is pivotal in maintaining genomic integrity. Pathogenic variation (PV) in DDR genes impairs their function, leading to genome instability and increased susceptibility to diseases, especially cancer. Understanding the evolution origin and arising time of DDR PV is crucial for comprehending disease susceptibility in modern humans.
Methods: We used big data approach to identify the PVs in DDR genes in modern humans. We mined multiple genomic databases derived from 251,214 modern humans of African and non-Africans. We compared the DDR PVs between African and non-African. We also mined the DDR PVs in the genomic data derived from 5,031 ancient humans. We used the DDR PVs from ancient humans as the intermediate to further the DDR PVs between African and non-African.
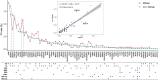
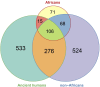
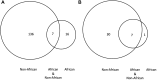
Results and discussion: We identified 1,060 single-base DDR PVs across 77 DDR genes in modern humans of African and non-African. Direct comparison of the DDR PVs between African and non-African showed that 82.1% of the non-African PVs were not present in African. We further identified 397 single-base DDR PVs in 56 DDR genes in the 5,031 ancient humans dated between 45,045 and 100 years before present (BP) lived in Eurasian continent therefore the descendants of the latest out-of-Africa human migrants occurred 50,000-60,000 years ago. By referring to the ancient DDR PVs, we observed that 276 of the 397 (70.3%) ancient DDR PVs were exclusive in non-African, 106 (26.7%) were shared between non-African and African, and only 15 (3.8%) were exclusive in African. We further validated the distribution pattern by testing the PVs in BRCA and TP53, two of the important genes in genome stability maintenance, in African, non-African, and Ancient humans. Our study revealed that DDR PVs in modern humans mostly emerged after the latest out-of-Africa migration. The data provides a foundation to understand the evolutionary basis of disease susceptibility, in particular cancer, in modern humans.
Keywords: BRCA; DDR genes; TP53; evolution origin; pathogenic variants.
Copyright © 2024 He, Kou, Li, Ding and Wang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Pathogenic variants in human DNA damage repair genes mostly arose in recent human history.BMC Cancer. 2024 Apr 4;24(1):415. doi: 10.1186/s12885-024-12160-6. BMC Cancer. 2024. PMID: 38575974 Free PMC article.
-
Using Portuguese BRCA pathogenic variation as a model to study the impact of human admixture on human health.BMC Genomics. 2024 Apr 27;25(1):416. doi: 10.1186/s12864-024-10311-4. BMC Genomics. 2024. PMID: 38671360 Free PMC article.
-
Evolutionary origin of germline pathogenic variants in human DNA mismatch repair genes.Hum Genomics. 2024 Jan 29;18(1):5. doi: 10.1186/s40246-024-00573-0. Hum Genomics. 2024. PMID: 38287404 Free PMC article.
-
Li-Fraumeni syndrome: not a straightforward diagnosis anymore-the interpretation of pathogenic variants of low allele frequency and the differences between germline PVs, mosaicism, and clonal hematopoiesis.Breast Cancer Res. 2019 Sep 18;21(1):107. doi: 10.1186/s13058-019-1193-1. Breast Cancer Res. 2019. PMID: 31533767 Free PMC article. Review.
-
Beyond BRCA: The Emerging Significance of DNA Damage Response and Personalized Treatment in Pancreatic and Prostate Cancer Patients.Int J Mol Sci. 2022 Apr 24;23(9):4709. doi: 10.3390/ijms23094709. Int J Mol Sci. 2022. PMID: 35563100 Free PMC article. Review.
References
-
- Bhaskaran S. P., Huang T., Rajendran B. K., Guo M., Luo J., Qin Z., et al. (2021). Ethnic-specific BRCA1/2 variation within Asia population: evidence from over 78 000 cancer and 40 000 non-cancer cases of Indian, Chinese, Korean and Japanese populations. J. Med. Genet. 58 (11), 752–759. 10.1136/jmedgenet-2020-107299 - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous


