Sequential glycosylations at the multibasic cleavage site of SARS-CoV-2 spike protein regulate viral activity
- PMID: 38755139
- PMCID: PMC11099032
- DOI: 10.1038/s41467-024-48503-x
Sequential glycosylations at the multibasic cleavage site of SARS-CoV-2 spike protein regulate viral activity
Abstract
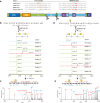
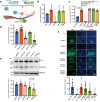
The multibasic furin cleavage site at the S1/S2 boundary of the spike protein is a hallmark of SARS-CoV-2 and plays a crucial role in viral infection. However, the mechanism underlying furin activation and its regulation remain poorly understood. Here, we show that GalNAc-T3 and T7 jointly initiate clustered O-glycosylations in the furin cleavage site of the SARS-CoV-2 spike protein, which inhibit furin processing, suppress the incorporation of the spike protein into virus-like-particles and affect viral infection. Mechanistic analysis reveals that the assembly of the spike protein into virus-like particles relies on interactions between the furin-cleaved spike protein and the membrane protein of SARS-CoV-2, suggesting a possible mechanism for furin activation. Interestingly, mutations in the spike protein of the alpha and delta variants of the virus confer resistance against glycosylation by GalNAc-T3 and T7. In the omicron variant, additional mutations reverse this resistance, making the spike protein susceptible to glycosylation in vitro and sensitive to GalNAc-T3 and T7 expression in human lung cells. Our findings highlight the role of glycosylation as a defense mechanism employed by host cells against SARS-CoV-2 and shed light on the evolutionary interplay between the host and the virus.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interest.
Figures
Similar articles
-
Rapid SARS-CoV-2 Adaptation to Available Cellular Proteases.J Virol. 2022 Mar 9;96(5):e0218621. doi: 10.1128/jvi.02186-21. Epub 2022 Jan 12. J Virol. 2022. PMID: 35019723 Free PMC article.
-
Furin cleavage of the SARS-CoV-2 spike is modulated by O-glycosylation.Proc Natl Acad Sci U S A. 2021 Nov 23;118(47):e2109905118. doi: 10.1073/pnas.2109905118. Proc Natl Acad Sci U S A. 2021. PMID: 34732583 Free PMC article.
-
A Multibasic Cleavage Site in the Spike Protein of SARS-CoV-2 Is Essential for Infection of Human Lung Cells.Mol Cell. 2020 May 21;78(4):779-784.e5. doi: 10.1016/j.molcel.2020.04.022. Epub 2020 May 1. Mol Cell. 2020. PMID: 32362314 Free PMC article.
-
Why All the Fury over Furin?J Med Chem. 2022 Feb 24;65(4):2747-2784. doi: 10.1021/acs.jmedchem.1c00518. Epub 2021 Aug 2. J Med Chem. 2022. PMID: 34340303 Review.
-
Roles of the polybasic furin cleavage site of spike protein in SARS-CoV-2 replication, pathogenesis, and host immune responses and vaccination.J Med Virol. 2022 May;94(5):1815-1820. doi: 10.1002/jmv.27539. Epub 2021 Dec 31. J Med Virol. 2022. PMID: 34936124 Review.
Cited by
-
O-glycosylation of SARS-CoV-2 spike protein by host O-glycosyltransferase strengthens its trimeric structure.Acta Biochim Biophys Sin (Shanghai). 2024 Jul 26;56(8):1118-1129. doi: 10.3724/abbs.2024127. Acta Biochim Biophys Sin (Shanghai). 2024. PMID: 39066577 Free PMC article.
References
MeSH terms
Substances
Supplementary concepts
Grants and funding
- 2021YFA1200903/Ministry of Science and Technology of the People's Republic of China (Chinese Ministry of Science and Technology)
- 91853131/National Natural Science Foundation of China (National Science Foundation of China)
- 81872786/National Natural Science Foundation of China (National Science Foundation of China)
- 202103000029/Guangzhou Science and Technology Program key projects
- 2021B1515130005/Guangdong Science and Technology Department (Science and Technology Department, Guangdong Province)
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous


