Classification of MLH1 Missense VUS Using Protein Structure-Based Deep Learning-Ramachandran Plot-Molecular Dynamics Simulations Method
- PMID: 38255924
- PMCID: PMC10815254
- DOI: 10.3390/ijms25020850
Classification of MLH1 Missense VUS Using Protein Structure-Based Deep Learning-Ramachandran Plot-Molecular Dynamics Simulations Method
Abstract
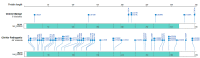
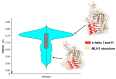
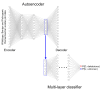
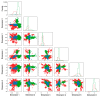
Pathogenic variation in DNA mismatch repair (MMR) gene MLH1 is associated with Lynch syndrome (LS), an autosomal dominant hereditary cancer. Of the 3798 MLH1 germline variants collected in the ClinVar database, 38.7% (1469) were missense variants, of which 81.6% (1199) were classified as Variants of Uncertain Significance (VUS) due to the lack of functional evidence. Further determination of the impact of VUS on MLH1 function is important for the VUS carriers to take preventive action. We recently developed a protein structure-based method named "Deep Learning-Ramachandran Plot-Molecular Dynamics Simulation (DL-RP-MDS)" to evaluate the deleteriousness of MLH1 missense VUS. The method extracts protein structural information by using the Ramachandran plot-molecular dynamics simulation (RP-MDS) method, then combines the variation data with an unsupervised learning model composed of auto-encoder and neural network classifier to identify the variants causing significant change in protein structure. In this report, we applied the method to classify 447 MLH1 missense VUS. We predicted 126/447 (28.2%) MLH1 missense VUS were deleterious. Our study demonstrates that DL-RP-MDS is able to classify the missense VUS based solely on their impact on protein structure.
Keywords: MLH1; Ramachandran plot; VUS; autoencoder; deep learning; molecular dynamics simulation; neural network.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Methylation Tolerance-Based Functional Assay to Assess Variants of Unknown Significance in the MLH1 and MSH2 Genes and Identify Patients With Lynch Syndrome.Gastroenterology. 2019 Aug;157(2):421-431. doi: 10.1053/j.gastro.2019.03.071. Epub 2019 Apr 15. Gastroenterology. 2019. PMID: 30998989
-
Three-step site-directed mutagenesis screen identifies pathogenic MLH1 variants associated with Lynch syndrome.J Med Genet. 2020 May;57(5):308-315. doi: 10.1136/jmedgenet-2019-106520. Epub 2019 Nov 29. J Med Genet. 2020. PMID: 31784484
-
Germline MLH1, MSH2 and MSH6 variants in Brazilian patients with colorectal cancer and clinical features suggestive of Lynch Syndrome.Cancer Med. 2018 May;7(5):2078-2088. doi: 10.1002/cam4.1316. Epub 2018 Mar 25. Cancer Med. 2018. PMID: 29575718 Free PMC article.
-
Identification of Lynch Syndrome.Gastrointest Endosc Clin N Am. 2022 Jan;32(1):45-58. doi: 10.1016/j.giec.2021.09.002. Gastrointest Endosc Clin N Am. 2022. PMID: 34798986 Review.
-
Correlation of tumour BRAF mutations and MLH1 methylation with germline mismatch repair (MMR) gene mutation status: a literature review assessing utility of tumour features for MMR variant classification.J Med Genet. 2012 Mar;49(3):151-7. doi: 10.1136/jmedgenet-2011-100714. J Med Genet. 2012. PMID: 22368298 Review.
Cited by
-
Special Issue: "Molecular Dynamics Simulations and Structural Analysis of Protein Domains".Int J Mol Sci. 2024 Oct 8;25(19):10793. doi: 10.3390/ijms251910793. Int J Mol Sci. 2024. PMID: 39409122 Free PMC article.
-
In Silico Deciphering of the Potential Impact of Variants of Uncertain Significance in Hereditary Colorectal Cancer Syndromes.Cells. 2024 Aug 6;13(16):1314. doi: 10.3390/cells13161314. Cells. 2024. PMID: 39195204 Free PMC article. Review.
References
-
- Dai J., Sanchez A., Adam C., Ranjha L., Reginato G., Chervy P., Tellier-Lebegue C., Andreani J., Guérois R., Ropars V., et al. Molecular basis of the dual role of the Mlh1-Mlh3 endonuclease in MMR and in meiotic crossover formation. Proc. Natl. Acad. Sci. USA. 2021;118:e2022704118. doi: 10.1073/pnas.2022704118. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- 085/2017/A2/the Macau Science and Technology Development Fund
- 0077/2019/AMJ/the Macau Science and Technology Development Fund
- 0032-2022-A1/the Macau Science and Technology Development Fund
- SRG2017-00097-FHS/The University of Macau
- MYRG2019-00018-FHS/The University of Macau
- 2020-00094-FHS/The University of Macau
- Startup fund/The Faculty of Health Sciences of University of Macau
- FHSIG/SW/0007/2020P/The Faculty of Health Sciences of University of Macau
- FHS Innovation grant/The Faculty of Health Sciences of University of Macau
- MYRG2022-00023-FHS/the University of Macau
- UMMTP-FDCT/0027/APD/2021/Postdoctoral Fellowship of Macau Science and Technology Development Fund
- FHS-CC-041-001-2023/The Faculty of Health Sciences of University of Macau
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous


